1CD5
 
 | | GLUCOSAMINE-6-PHOSPHATE DEAMINASE FROM E.COLI, T CONFORMER | | Descriptor: | PROTEIN (GLUCOSAMINE 6-PHOSPHATE DEAMINASE) | | Authors: | Horjales, E, Altamirano, M.M, Calcagno, M.L, Garratt, R.C, Oliva, G. | | Deposit date: | 1999-03-05 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric transition of glucosamine-6-phosphate deaminase: the structure of the T state at 2.3 A resolution.
Structure Fold.Des., 7, 1999
|
|
4WCH
 
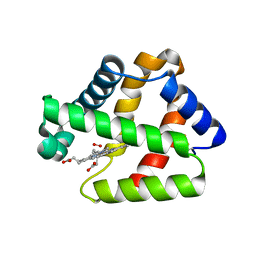 | | Structure of Isolated D Chain of Gigant Hemoglobin from Glossoscolex paulistus | | Descriptor: | Isolated Chain D of Gigant Hemoglobin from Glossoscolex Paulistus, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bachega, J.F.R, Maluf, F.V, Pereira, H.M, Brandao-Neto, J, Tabak, M, Garratt, R.C, Horjales, E. | | Deposit date: | 2014-09-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of the giant haemoglobin from Glossoscolex paulistus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1HOT
 
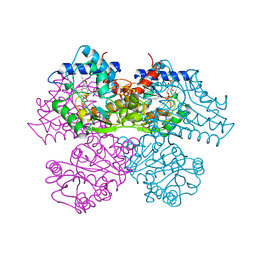 | | GLUCOSAMINE 6-PHOSPHATE DEAMINASE COMPLEXED WITH THE ALLOSTERIC ACTIVATOR N-ACETYL-GLUCOSAMINE-6-PHOSPHATE | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.L, Garratt, R, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-11-17 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
4U8U
 
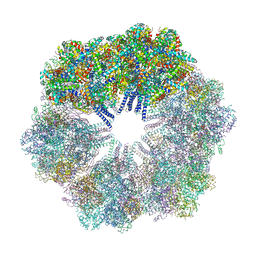 | | The Crystallographic structure of the giant hemoglobin from Glossoscolex paulistus at 3.2 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Bachega, J.F.R, Maluf, F.V, Andi, B, D'Muniz Pereira, H, Carazzollea, M.F, Orville, A, Tabak, M, Garratt, R.C, Horjales, E. | | Deposit date: | 2014-08-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the giant haemoglobin from Glossoscolex paulistus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1NE7
 
 | | HUMAN GLUCOSAMINE-6-PHOSPHATE DEAMINASE ISOMERASE AT 1.75 A RESOLUTION COMPLEXED WITH N-ACETYL-GLUCOSAMINE-6-PHOSPHATE AND 2-DEOXY-2-AMINO-GLUCITOL-6-PHOSPHATE | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine-6-phosphate isomerase, ... | | Authors: | Arreola, R, Valderrama, B, Morante, M.L, Horjales, E. | | Deposit date: | 2002-12-10 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two mammalian glucosamine-6-phosphate deaminases: a structural and genetic study.
Febs Lett., 551, 2003
|
|
1SY7
 
 | | Crystal structure of the catalase-1 from Neurospora crassa, native structure at 1.75A resolution. | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Diaz, A, Horjales, E, Rudino-Pinera, E, Arreola, R, Hansberg, W. | | Deposit date: | 2004-04-01 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unusual Cys-Tyr covalent bond in a large catalase
J.Mol.Biol., 342, 2004
|
|
3EJ6
 
 | | Neurospora Crassa Catalase-3 Crystal Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Catalase-3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Diaz, A, Valdes, V.-J, Rudino-Pinera, E, Horjales, E, Hansberg, W. | | Deposit date: | 2008-09-17 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Function Relationships in Fungal Large-Subunit Catalases
J.Mol.Biol., 386, 2008
|
|
2WSR
 
 | | MONOTIM MUTANT RMM0-1, MONOMERIC FORM. | | Descriptor: | AZIDE ION, SULFATE ION, TRIOSE PHOSPHATE ISOMERASE, ... | | Authors: | Rudino-Pinera, E, Rojas-Trejo, S.P, Arreola, R, Saab-Rincon, G, Soberon, X, Horjales, E. | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Space Group Transition Driven by Temperature and Related to Monomer-Dimer Transition in Solution: The Case of Monomeric Tim of Trypanosoma Brucei Brucei
To be Published
|
|
1JT9
 
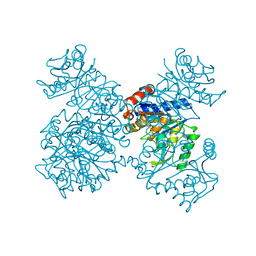 | | Structure of the mutant F174A T form of the Glucosamine-6-Phosphate deaminase from E.coli | | Descriptor: | Glucosamine-6-Phosphate deaminase | | Authors: | Bustos-Jaimes, I, Sosa-Peinado, A, Rudino-Pinera, E, Horjales, E, Calcagno, M.L. | | Deposit date: | 2001-08-20 | | Release date: | 2002-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | On the role of the conformational flexibility of the active-site lid on the allosteric kinetics of glucosamine-6-phosphate deaminase.
J.Mol.Biol., 319, 2002
|
|
1HOR
 
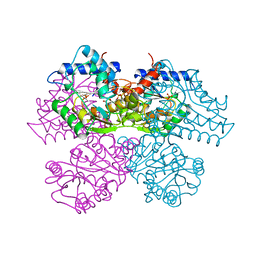 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
5V5T
 
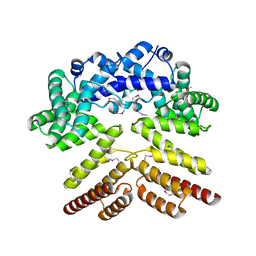 | | Crystal structure of leucine-rich protein regulator, ElrR, from Enterococcus faecalis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Conserved domain protein | | Authors: | De Groote, M.C.R, Camargo, I.L, Serror, P, Horjales, E. | | Deposit date: | 2017-03-15 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and functional characterization of ElrR, a member of the RRNPP family of transcriptional regulators.
To be published
|
|
5V5U
 
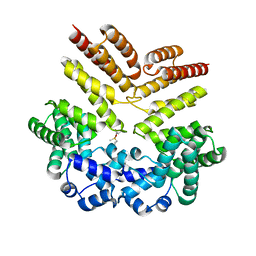 | | Crystal structure of leucine-rich protein regulator, ElrR, from Enterococcus faecalis | | Descriptor: | Conserved domain protein | | Authors: | De Groote, M.C.R, Camargo, I.L, Serror, P, Horjales, E. | | Deposit date: | 2017-03-15 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | Crystal structure of leucine-rich protein regulator, ElrR, from Enterococcus faecalis
To be published
|
|
1FRZ
 
 | | GLUCOSAMINE-6-PHOSPHATE DEAMINASE FROM E.COLI, R CONFORMER. COMPLEXED WITH THE ALLOSTERIC ACTIVATOR N-ACETYL-GLUCOSAMINE-6-PHOSPHATE AT 2.2 A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rudino-Pinera, E, Morales-Arrieta, S, Rojas-Trejo, S.P, Horjales, E. | | Deposit date: | 2000-09-07 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural flexibility, an essential component of the allosteric activation in Escherichia coli glucosamine-6-phosphate deaminase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1FSF
 
 | | GLUCOSAMINE-6-PHOSPHATE DEAMINASE FROM E.COLI, T CONFORMER, AT 1.9A RESOLUTION | | Descriptor: | GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rudino-Pinera, E, Morales-Arrieta, S, Rojas-Trejo, S.P, Horjales, E. | | Deposit date: | 2000-09-08 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility, an essential component of the allosteric activation in Escherichia coli glucosamine-6-phosphate deaminase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1FQO
 
 | | GLUCOSAMINE 6-PHOSPHATE DEAMINASE COMPLEXED WITH THE SUBSTRATE OF THE REVERSE REACTION FRUCTOSE 6-PHOSPHATE (OPEN FORM) | | Descriptor: | FRUCTOSE -6-PHOSPHATE, GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rudino-Pinera, E, Morales-Arrieta, S, Rojas-Trejo, S.P, Horjales, E. | | Deposit date: | 2000-09-06 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural flexibility, an essential component of the allosteric activation in Escherichia coli glucosamine-6-phosphate deaminase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1FS5
 
 | | A DISCOVERY OF THREE ALTERNATE CONFORMATIONS IN THE ACTIVE SITE OF GLUCOSAMINE-6-PHOSPHATE ISOMERASE | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE-6-PHOSPHATE DEAMINASE, L(+)-TARTARIC ACID | | Authors: | Rudino-Pinera, E, Morales-Arrieta, S, Rojas-Trejo, S.P, Horjales, E. | | Deposit date: | 2000-09-08 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural flexibility, an essential component of the allosteric activation in Escherichia coli glucosamine-6-phosphate deaminase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1FS6
 
 | | GLUCOSAMINE-6-PHOSPHATE DEAMINASE FROM E.COLI, T CONFORMER, AT 2.2A RESOLUTION | | Descriptor: | GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rudino-Pinera, E, Morales-Arrieta, S, Rojas-Trejo, S.P, Horjales, E. | | Deposit date: | 2000-09-08 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural flexibility, an essential component of the allosteric activation in Escherichia coli glucosamine-6-phosphate deaminase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1DEA
 
 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
2WME
 
 | | Crystallographic structure of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | BETA-MERCAPTOETHANOL, BETAINE ALDEHYDE DEHYDROGENASE, GLYCEROL, ... | | Authors: | Gonzalez-Segura, L, Rudino-Pinera, E, Munoz-Clares, R.A, Horjales, E. | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a Ternary Complex of Betaine Aldehyde Dehydrogenase from Pseudomonas Aeruginosa Provides New Insight Into the Reaction Mechanism and Shows a Novel Binding Mode of the 2'- Phosphate of Nadp(+) and a Novel Cation Binding Site.
J.Mol.Biol., 385, 2009
|
|
2WU1
 
 | |
2W0L
 
 | |
2W0K
 
 | |
2WSQ
 
 | | MonoTIM mutant RMM0-1, dimeric form. | | Descriptor: | SULFATE ION, TRIOSE PHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Rudino-Pinera, E, Rojas-Trejo, S.P, Arreola, R, Saab-Rincon, G, Soberon, X, Horjales, E. | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Space Group Transition Driven by Temperature and Related to Monomer-Dimer Transition in Solution: The Case of Monomeric Tim of Trypanosoma Brucei Brucei
To be Published
|
|
4A2H
 
 | | Crystal Structure of Laccase from Coriolopsis gallica pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | De La Mora, E, Valderrama, B, Horjales, E, Rudino-Pinera, E. | | Deposit date: | 2011-09-27 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Changes Caused by Radiation-Induced Reduction and Radiolysis: The Effect of X-Ray Absorbed Dose in a Fungal Multicopper Oxidase
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4A2D
 
 | | Coriolopsis gallica Laccase T2 Copper Depleted at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, LACCASE, ... | | Authors: | De La Mora, E, Valderrama, B, Horjales, E, Rudino-Pinera, E. | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Changes Caused by Radiation-Induced Reduction and Radiolysis: The Effect of X-Ray Absorbed Dose in a Fungal Multicopper Oxidase
Acta Crystallogr.,Sect.D, 68, 2012
|
|
