1U8R
 
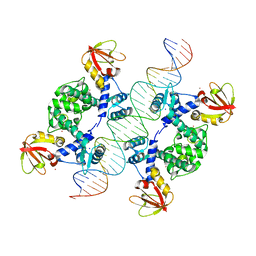 | | Crystal Structure of an IdeR-DNA Complex Reveals a Conformational Change in Activated IdeR for Base-specific Interactions | | Descriptor: | COBALT (II) ION, Iron-dependent repressor ideR, SODIUM ION, ... | | Authors: | Wisedchaisri, G, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of an IdeR-DNA Complex Reveals a Conformational Change in Activated IdeR for Base-specific Interactions.
J.Mol.Biol., 342, 2004
|
|
2A0S
 
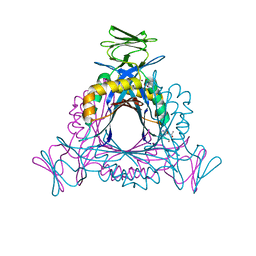 | |
2BH1
 
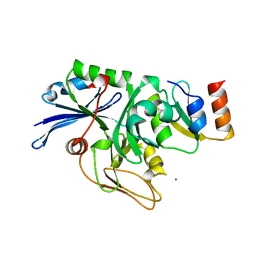 | | X-ray structure of the general secretion pathway complex of the N- terminal domain of EpsE and the cytosolic domain of EpsL of Vibrio cholerae | | Descriptor: | CALCIUM ION, GENERAL SECRETION PATHWAY PROTEIN E,, GENERAL SECRETION PATHWAY PROTEIN L | | Authors: | Abendroth, J, Murphy, P.M, Mushtaq, A, Bagdasarian, M, Sandkvist, M, Hol, W.G.J. | | Deposit date: | 2005-01-06 | | Release date: | 2005-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-Ray Structure of the Type II Secretion System Complex Formed by the N-Terminal Domain of Epse and the Cytoplasmic Domain of Epsl of Vibrio Cholerae
J.Mol.Biol., 348, 2005
|
|
2BTM
 
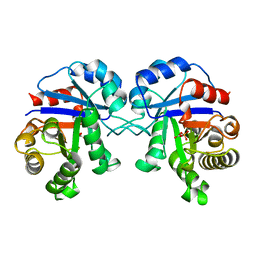 | |
2AF6
 
 | | Crystal structure of Mycobacterium tuberculosis Flavin dependent thymidylate synthase (Mtb ThyX) in the presence of co-factor FAD and substrate analog 5-Bromo-2'-Deoxyuridine-5'-Monophosphate (BrdUMP) | | Descriptor: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Sampathkumar, P, Turley, S, Ulmer, J.E, Rhie, H.G, Sibley, C.H, Hol, W.G. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the Mycobacterium tuberculosis Flavin Dependent Thymidylate Synthase (MtbThyX) at 2.0A Resolution.
J.Mol.Biol., 352, 2005
|
|
2B94
 
 | |
2A0K
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.8 A resolution | | Descriptor: | GLYCEROL, Nucleoside 2-deoxyribosyltransferase, SULFATE ION | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-06-16 | | Release date: | 2005-07-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2A0U
 
 | |
2B51
 
 | | Structural Basis for UTP Specificity of RNA Editing TUTases from Trypanosoma Brucei | | Descriptor: | MANGANESE (II) ION, RNA editing complex protein MP57, URIDINE 5'-TRIPHOSPHATE | | Authors: | Deng, J, Ernst, N.L, Turley, S, Stuart, K.D, Hol, W.G. | | Deposit date: | 2005-09-27 | | Release date: | 2005-11-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei.
Embo J., 24, 2005
|
|
1ZLJ
 
 | | Crystal Structure of the Mycobacterium tuberculosis Hypoxic Response Regulator DosR C-terminal Domain | | Descriptor: | Dormancy Survival Regulator | | Authors: | Wisedchaisri, G, Wu, M, Rice, A.E, Roberts, D.M, Sherman, D.R, Hol, W.G.J. | | Deposit date: | 2005-05-06 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Mycobacterium tuberculosis DosR and DosR-DNA complex involved in gene activation during adaptation to hypoxic latency.
J.Mol.Biol., 354, 2005
|
|
1TII
 
 | |
2B30
 
 | |
2B4V
 
 | | Structural Basis for UTP Specificity of RNA Editing TUTases From Trypanosoma Brucei | | Descriptor: | POTASSIUM ION, RNA editing complex protein MP57 | | Authors: | Deng, J, Ernst, N.L, Turley, S, Stuart, K.D, Hol, W.G. | | Deposit date: | 2005-09-26 | | Release date: | 2005-11-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei.
Embo J., 24, 2005
|
|
2B56
 
 | | Structural Basis for UTP Specificity of RNA Editing TUTases From Trypanosoma Brucei | | Descriptor: | MAGNESIUM ION, RNA editing complex protein MP57, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Deng, J, Ernst, N.L, Turley, S, Stuart, K.D, Hol, W.G. | | Deposit date: | 2005-09-27 | | Release date: | 2005-11-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei.
Embo J., 24, 2005
|
|
2DTR
 
 | | STRUCTURE OF DIPHTHERIA TOXIN REPRESSOR | | Descriptor: | COBALT (II) ION, DIPHTHERIA TOXIN REPRESSOR, SULFATE ION | | Authors: | Qiu, X, Pohl, E, Hol, W.G. | | Deposit date: | 1996-07-26 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of the diphtheria toxin repressor complexed with cobalt and manganese reveals an SH3-like third domain and suggests a possible role of phosphate as co-corepressor.
Biochemistry, 35, 1996
|
|
1LT4
 
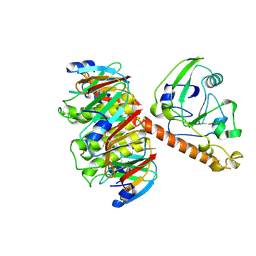 | | HEAT-LABILE ENTEROTOXIN MUTANT S63K | | Descriptor: | HEAT-LABILE ENTEROTOXIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Van Den Akker, F, Hol, W.G.J. | | Deposit date: | 1997-04-14 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a non-toxic mutant of heat-labile enterotoxin, which is a potent mucosal adjuvant.
Protein Sci., 6, 1997
|
|
1QCB
 
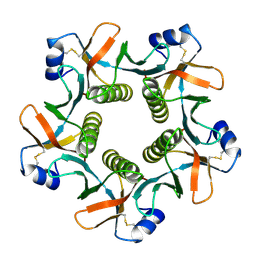 | |
1DPB
 
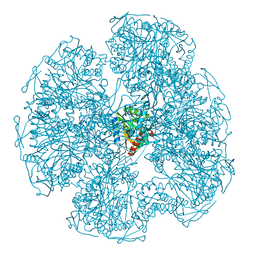 | |
1QB5
 
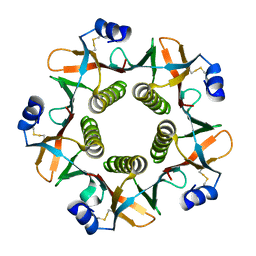 | |
1EAF
 
 | |
1QS0
 
 | | Crystal Structure of Pseudomonas Putida 2-oxoisovalerate Dehydrogenase (Branched-Chain Alpha-Keto Acid Dehydrogenase, E1B) | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, 2-OXOISOVALERATE DEHYDROGENASE ALPHA-SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA-SUBUNIT, ... | | Authors: | Aevarsson, A, Seger, K, Turley, S, Sokatch, J.R, Hol, W.G.J. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-18 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of 2-oxoisovalerate and dehydrogenase and the architecture of 2-oxo acid dehydrogenase multienzyme complexes.
Nat.Struct.Biol., 6, 1999
|
|
1EFI
 
 | |
1DPD
 
 | |
1R49
 
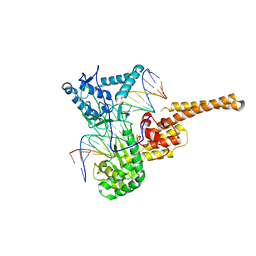 | | Human topoisomerase I (Topo70) double mutant K532R/Y723F | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(P*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3', DNA topoisomerase I | | Authors: | Interthal, H, Quigley, P.M, Hol, W.G, Champoux, J.J. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | The role of lysine 532 in the catalytic mechanism of human topoisomerase I.
J.Biol.Chem., 279, 2004
|
|
1DPC
 
 | |
