2DDH
 
 | | Crystal Structure of Acyl-CoA oxidase complexed with 3-OH-dodecanoate | | Descriptor: | (3R)-3-HYDROXYDODECANOIC ACID, Acyl-CoA oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Keiji, T, Nakajima, Y, Miyahara, I, Hirotsu, K. | | Deposit date: | 2006-01-29 | | Release date: | 2006-03-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Three-Dimensional Structure of Rat-Liver Acyl-CoA Oxidase in Complex with a Fatty Acid: Insights into Substrate-Recognition and Reactivity toward Molecular Oxygen.
J.Biochem.(Tokyo), 139, 2006
|
|
1WKH
 
 | | Acetylornithine aminotransferase from thermus thermophilus HB8 | | Descriptor: | 4-[(1,3-DICARBOXY-PROPYLAMINO)-METHYL]-3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDINIUM, Acetylornithine/acetyl-lysine aminotransferase | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Acetylornithine aminotransferase from thermus thermophilus HB8
To be Published
|
|
1WKG
 
 | | Acetylornithine aminotransferase from thermus thermophilus HB8 | | Descriptor: | Acetylornithine/acetyl-lysine aminotransferase, N~2~-ACETYL-N~5~-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-ORNITHINE | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Acetylornithine aminotransferase from thermus thermophilus HB8
To be Published
|
|
1WTC
 
 | | Crystal Structure of S.pombe Serine Racemase complex with AMPPCP | | Descriptor: | Hypothetical protein C320.14 in chromosome III, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Goto, M, Miyahara, I, Hirotsu, K. | | Deposit date: | 2004-11-22 | | Release date: | 2005-11-01 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a homolog of mammalian serine racemase from Schizosaccharomyces pombe
J.Biol.Chem., 284, 2009
|
|
1JMX
 
 | |
1I1L
 
 | | CRYSTAL STRUCTURE OF ESCHELICHIA COLI BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE. | | Descriptor: | 2-METHYLLEUCINE, BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okada, K, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2001-02-02 | | Release date: | 2001-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Escherichia coli branched-chain amino acid aminotransferase and its complexes with 4-methylvalerate and 2-methylleucine: induced fit and substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
1I1M
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE. | | Descriptor: | 4-METHYL VALERIC ACID, BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okada, K, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2001-02-02 | | Release date: | 2001-07-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Escherichia coli branched-chain amino acid aminotransferase and its complexes with 4-methylvalerate and 2-methylleucine: induced fit and substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
1VEF
 
 | | Acetylornithine aminotransferase from Thermus thermophilus HB8 | | Descriptor: | Acetylornithine/acetyl-lysine aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-30 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Three-Dimensional Strutcure of Acetylornithine aminotransferase from Thermus thermophilus HB8
To be Published
|
|
1JMZ
 
 | | crystal structure of a quinohemoprotein amine dehydrogenase from pseudomonas putida with inhibitor | | Descriptor: | Amine Dehydrogenase, HEME C, NICKEL (II) ION, ... | | Authors: | Satoh, A, Miyahara, I, Hirotsu, K. | | Deposit date: | 2001-07-20 | | Release date: | 2002-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of quinohemoprotein amine dehydrogenase from Pseudomonas putida. Identification of a novel quinone cofactor encaged by multiple thioether cross-bridges.
J.Biol.Chem., 277, 2002
|
|
2DKJ
 
 | | Crystal Structure of T.th.HB8 Serine Hydroxymethyltransferase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, serine hydroxymethyltransferase | | Authors: | Kai, K, Goto, M, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-11 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of T.th.HB8 Serine Hydroxymethyltransferase
To be Published
|
|
2YRR
 
 | | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8 | | Descriptor: | Aminotransferase, class V, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Miyahara, I, Matsumura, M, Goto, M, Omi, R, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | hypothetical alanine aminotransferase (TTH0173) from Thermus thermophilus HB8
To be Published
|
|
2YRI
 
 | | Crystal structure of alanine-pyruvate aminotransferase with 2-methylserine | | Descriptor: | (S,E)-3-HYDROXY-2-((3-HYDROXY-2-METHYL-5-(PHOSPHONOOXYMETHYL)PYRIDIN-4-YL)METHYLENEAMINO)-2-METHYLPROPANOIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase, ... | | Authors: | Miyahara, I, Matsumura, M, Goto, M, Omi, R, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | hypothetical alanine aminotransferase (TTHA0173) from Thermus thermophilus HB8
To be Published
|
|
1IS2
 
 | |
1I1K
 
 | | CRYSTAL STRUCTURE OF ESCHELICHIA COLI BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE. | | Descriptor: | BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Okada, K, Hirotsu, K, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2001-02-02 | | Release date: | 2001-07-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Escherichia coli branched-chain amino acid aminotransferase and its complexes with 4-methylvalerate and 2-methylleucine: induced fit and substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
3A9Z
 
 | |
3A9Y
 
 | |
3A9X
 
 | | Crystal structure of rat selenocysteine lyase | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Selenocysteine lyase | | Authors: | Omi, R, Hirotsu, K. | | Deposit date: | 2009-11-08 | | Release date: | 2010-03-16 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction mechanism and molecular basis for selenium/sulfur discrimination of selenocysteine lyase.
J.Biol.Chem., 285, 2010
|
|
1AY8
 
 | |
1J21
 
 | |
1J1Z
 
 | |
1CQ8
 
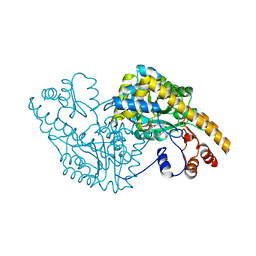 | | ASPARTATE AMINOTRANSFERASE (E.C. 2.6.1.1) COMPLEXED WITH C6-PYRIDOXAL-5P-PHOSPHATE | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO-HEXANOIC ACID, ASPARTATE AMINOTRANSFERASE (2.6.1.1) | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-06 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
1C9C
 
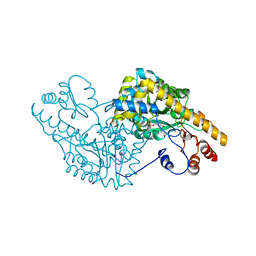 | | ASPARTATE AMINOTRANSFERASE COMPLEXED WITH C3-PYRIDOXAL-5'-PHOSPHATE | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
2CWH
 
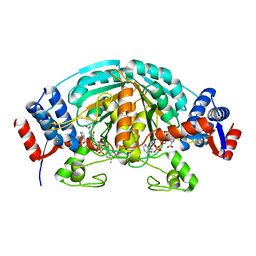 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae complexed with NADPH and pyrrole-2-carboxylate | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRROLE-2-CARBOXYLATE, delta1-piperideine-2-carboxylate reductase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2005-06-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
2CWF
 
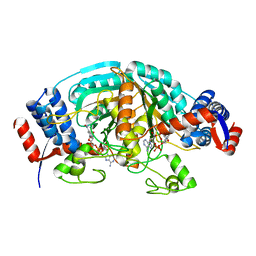 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, delta1-piperideine-2-carboxylate reductase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2005-06-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
1B5O
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE SINGLE MUTANT 1 | | Descriptor: | PHOSPHATE ION, PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Kawaguchi, S.I, Miyahara, I, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-01-07 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme
J.BIOCHEM.(TOKYO), 130, 2001
|
|
