1A0O
 
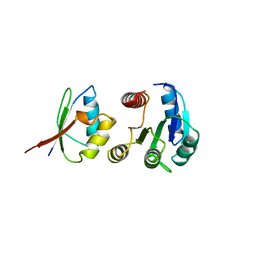 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY | | Descriptor: | CHEA, CHEY, MANGANESE (II) ION | | Authors: | Chinardet, N, Welch, M, Mourey, L, Birck, C, Samama, J.P. | | Deposit date: | 1997-12-05 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the CheY-binding domain of histidine kinase CheA in complex with CheY.
Nat.Struct.Biol., 5, 1998
|
|
3K0J
 
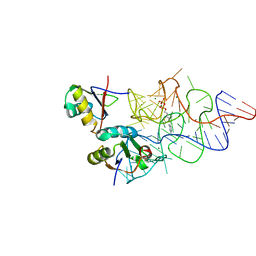 | | Crystal structure of the E. coli ThiM riboswitch in complex with thiamine pyrophosphate and the U1A crystallization module | | Descriptor: | MAGNESIUM ION, RNA (87-MER), THIAMINE DIPHOSPHATE, ... | | Authors: | Kulshina, N, Edwards, T.E, Ferre-D'Amare, A.R. | | Deposit date: | 2009-09-24 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch.
Rna, 16, 2010
|
|
3IWN
 
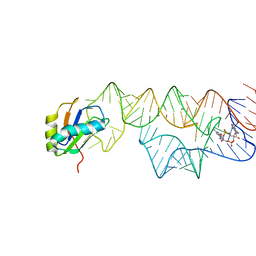 | | Co-crystal structure of a bacterial c-di-GMP riboswitch | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C-di-GMP riboswitch, U1 small nuclear ribonucleoprotein A | | Authors: | Kulshina, N, Baird, N.J, Ferre-D'Amare, A.R. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2TOD
 
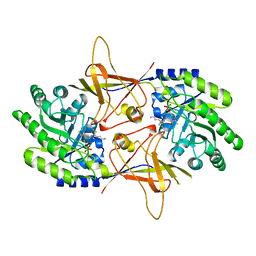 | | ORNITHINE DECARBOXYLASE FROM TRYPANOSOMA BRUCEI K69A MUTANT IN COMPLEX WITH ALPHA-DIFLUOROMETHYLORNITHINE | | Descriptor: | ALPHA-DIFLUOROMETHYLORNITHINE, PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Grishin, N.V, Osterman, A.L, Brooks, H.B, Phillips, M.A, Goldsmith, E.J. | | Deposit date: | 1999-05-18 | | Release date: | 1999-11-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine.
Biochemistry, 38, 1999
|
|
2WIM
 
 | | Crystal structure of NCAM2 IG1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAL CELL ADHESION MOLECULE 2 | | Authors: | Kulahin, N, Kristensen, O, Rasmussen, K, Kastrup, J, Berezin, V, Bock, E, Walmod, P, Gajhede, M. | | Deposit date: | 2009-05-13 | | Release date: | 2010-08-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural model and trans-interaction of the entire ectodomain of the olfactory cell adhesion molecule.
Structure, 19, 2011
|
|
2V5T
 
 | | Crystal structure of NCAM2 Ig2-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NEURAL CELL ADHESION MOLECULE 2, ... | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2007-07-10 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2XYC
 
 | | CRYSTAL STRUCTURE OF NCAM2 IGIV-FN3I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NEURAL CELL ADHESION MOLECULE 2, ... | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2010-11-17 | | Release date: | 2011-02-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2VAJ
 
 | | Crystal structure of NCAM2 Ig1 (I4122 cell unit) | | Descriptor: | NEURAL CELL ADHESION MOLECULE 2 | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Kastrup, J.S, Navarro-Poulsen, J.-C, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2007-08-31 | | Release date: | 2008-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal Structure of the Ig1 Domain of the Neural Cell Adhesion Molecule Ncam2 Displays Domain Swapping.
J.Mol.Biol., 382, 2008
|
|
2WFU
 
 | | Crystal structure of DILP5 variant DB | | Descriptor: | PROBABLE INSULIN-LIKE PEPTIDE 5 A CHAIN, PROBABLE INSULIN-LIKE PEPTIDE 5 B CHAIN | | Authors: | Kulahin, N, Schluckebier, G, Sajid, W, De Meyts, P. | | Deposit date: | 2009-04-15 | | Release date: | 2010-05-26 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biological Properties of the Drosophila Insulin-Like Peptide 5 Show Evolutionary Conservation.
J.Biol.Chem., 286, 2011
|
|
2WFV
 
 | | Crystal structure of DILP5 variant C4 | | Descriptor: | PROBABLE INSULIN-LIKE PEPTIDE 5 A CHAIN, PROBABLE INSULIN-LIKE PEPTIDE 5 B CHAIN | | Authors: | Kulahin, N, Schluckebier, G, Sajid, W, De Meyts, P. | | Deposit date: | 2009-04-15 | | Release date: | 2010-05-26 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biological Properties of the Drosophila Insulin-Like Peptide 5 Show Evolutionary Conservation.
J.Biol.Chem., 286, 2011
|
|
2UUS
 
 | | Crystal structure of the rat FGF1-sucrose octasulfate (SOS) complex. | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, HEPARIN-BINDING GROWTH FACTOR 1 | | Authors: | Kulahin, N, Kiselyov, V, Kochoyan, A, Kristensen, O, Berezin, V, Bock, E, Gajhede, M. | | Deposit date: | 2007-03-07 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dimerization Effect of Sucrose Octasulfate on Rat Fgf1.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4EFJ
 
 | |
4CBP
 
 | | Crystal structure of neural ectodermal development factor IMP-L2. | | Descriptor: | GLYCEROL, NEURAL/ECTODERMAL DEVELOPMENT FACTOR IMP-L2 | | Authors: | Kulahin, N, Kristensen, O, Brzozowski, M, Schluckebier, G, Meyts, P.D. | | Deposit date: | 2013-10-15 | | Release date: | 2014-10-29 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Analysis of Imp-L2 Function
To be Published
|
|
1QU4
 
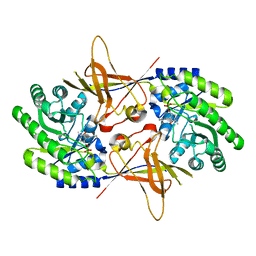 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE | | Descriptor: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Grishin, N.V, Osterman, A.L, Brooks, H.B, Phillips, M.A, Goldsmith, E.J. | | Deposit date: | 1999-07-06 | | Release date: | 1999-11-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine.
Biochemistry, 38, 1999
|
|
2XY1
 
 | | CRYSTAL STRUCTURE OF NCAM2 IG3-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEURAL CELL ADHESION MOLECULE 2 | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2XY2
 
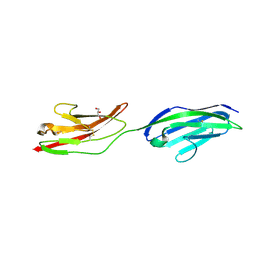 | | CRYSTAL STRUCTURE OF NCAM2 IG1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NEURAL CELL ADHESION MOLECULE 2 | | Authors: | Kulahin, N, Rasmussen, K.K, Kristensen, O, Berezin, V, Bock, E, Walmod, P.S, Gajhede, M. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2J3P
 
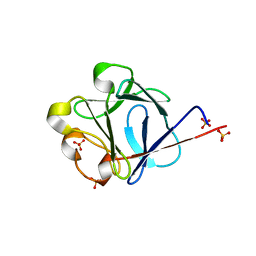 | | crystal structure of rat FGF1 at 1.4 A | | Descriptor: | HEPARIN-BINDING GROWTH FACTOR 1, SULFATE ION | | Authors: | Kulahin, N, Kristensen, O, Berezin, V, Gajhede, M, Bock, E. | | Deposit date: | 2006-08-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Rat Acidic Fibroblast Growth Factor at 1.4 A Resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2JLL
 
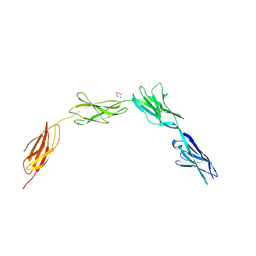 | | Crystal structure of NCAM2 IgIV-FN3II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Kulahin, N, Rasmussen, K, Kristensen, O, Kastrup, J, Berezin, V, Bock, E, Walmod, P, Gajhede, M. | | Deposit date: | 2008-09-10 | | Release date: | 2009-11-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
2ATH
 
 | | Crystal structure of the ligand binding domain of human PPAR-gamma im complex with an agonist | | Descriptor: | 2-{5-[3-(7-PROPYL-3-TRIFLUOROMETHYLBENZO[D]ISOXAZOL-6-YLOXY)PROPOXY]INDOL-1-YL}ETHANOIC ACID, Peroxisome proliferator activated receptor gamma | | Authors: | Mahindroo, N, Huang, C.-F, Wu, S.-Y, Hsieh, H.-P. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Novel indole-based peroxisome proliferator-activated receptor agonists: design, SAR, structural biology, and biological activities
J.Med.Chem., 48, 2005
|
|
5ZJ9
 
 | | human D-amino acid oxidase complexed with 5-chlorothiophene-3-carboxylic acid | | Descriptor: | 5-chloro thiophene-3-carboxylic acid, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kato, Y, Hin, N, Maita, N, Thomas, A.G, Kurosawa, S, Rojas, C, Yorita, K, Slusher, B.S, Fukui, K, Tsukamoto, T. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for potent inhibition of d-amino acid oxidase by thiophene carboxylic acids
Eur J Med Chem, 159, 2018
|
|
5ZJA
 
 | | human D-amino acid oxidase complexed with 5-chlorothiophene-2-carboxylic acid | | Descriptor: | 5-chloro thiophene-2-carboxylic acid, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kato, Y, Hin, N, Maita, N, Thomas, A.G, Kurosawa, S, Rojas, C, Yorita, K, Slusher, B.S, Fukui, K, Tsukamoto, T. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for potent inhibition of d-amino acid oxidase by thiophene carboxylic acids
Eur J Med Chem, 159, 2018
|
|
5VJX
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
5VJI
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-07 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
6WNV
 
 | | 70S ribosome without free 5S rRNA and with a perturbed PTC | | Descriptor: | 16S ribosomal RNA, 23s-5s joint ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A, Mankin, A.S, Huang, S, Aleksashin, N.A, Klepacki, D, Reier, K, Kefi, A, Szal, A, Remme, J, Jaeger, L, Vazquez-Laslop, N. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly.
Nat Commun, 11, 2020
|
|
1LW7
 
 | | NADR PROTEIN FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, TRANSCRIPTIONAL REGULATOR NADR | | Authors: | Singh, S.K, Kurnasov, O.V, Chen, B, Robinson, H, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-05-30 | | Release date: | 2002-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities.
J.Biol.Chem., 277, 2002
|
|
