1EG2
 
 | | CRYSTAL STRUCTURE OF RHODOBACTER SPHEROIDES (N6 ADENOSINE) METHYLTRANSFERASE (M.RSRI) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MODIFICATION METHYLASE RSRI | | Authors: | Scavetta, R.D, Thomas, C.B, Walsh, M.A, Szegedi, S, Joachimiak, A, Gumport, R.I, Churchill, M.E.A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-02-11 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases.
Nucleic Acids Res., 28, 2000
|
|
2CNG
 
 | | Structural Insights into the Design of Nonpeptidic Isothiazolidinone- Containing Inhibitors of Protein Tyrosine Phosphatase 1B | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-{(1S)-2-{4-[(5R)-1,1-DIOXIDO-3-OXOISOTHIAZOLIDIN-5-YL]PHENYL}-1-[5-(TRIFLUOROMETHYL)-1H-BENZIMIDAZOL-2-YL]ETHYL}-2,2,2-TRIFLUOROACETAMIDE, ... | | Authors: | Ala, P.J, Gonneville, L, Hillman, M, Becker-Pasha, M, Yue, E.W, Douty, B, Wayland, B, Polam, P, Crawley, M.L, McLaughlin, E, Sparks, R.B, Glass, B, Takvorian, A, Combs, A.P, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-21 | | Release date: | 2006-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights Into the Design of Nonpeptidic Isothiazolidinone-Containing Inhibitors of Protein- Tyrosine Phosphatase 1B.
J.Biol.Chem., 281, 2006
|
|
2A18
 
 | | carboxysome shell protein ccmK4, crystal form 2 | | Descriptor: | AMMONIUM ION, Carbon dioxide concentrating mechanism protein ccmK homolog 4 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-18 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
3QH9
 
 | | Human Liprin-beta2 Coiled-Coil | | Descriptor: | AMMONIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Stafford, R.L, Tang, M, Phillips, M.L, Bowie, J.U. | | Deposit date: | 2011-01-25 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the central coiled-coil domain from human liprin-beta2
Biochemistry, 50, 2011
|
|
1D7K
 
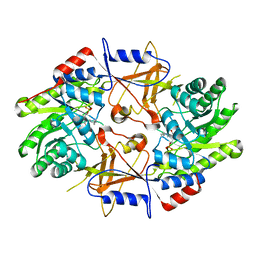 | | CRYSTAL STRUCTURE OF HUMAN ORNITHINE DECARBOXYLASE AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN ORNITHINE DECARBOXYLASE | | Authors: | Almrud, J.J, Oliveira, M.A, Kern, A.D, Grishin, N.V, Phillips, M.A, Hackert, M.L. | | Deposit date: | 1999-10-18 | | Release date: | 2000-10-25 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human ornithine decarboxylase at 2.1 A resolution: structural insights to antizyme binding.
J.Mol.Biol., 295, 2000
|
|
1CMQ
 
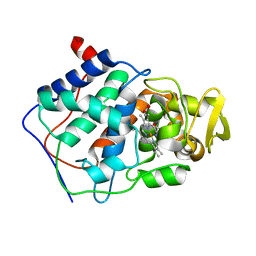 | | SMALL MOLECULE BINDING TO AN ARTIFICIALLY CREATED CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Mcree, D.E, Churchill, M.J, Goodin, D.B. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small molecule binding to an artificially created cavity at the active site of cytochrome c peroxidase.
Biochemistry, 33, 1994
|
|
2TOD
 
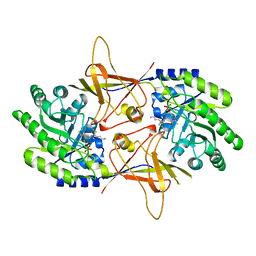 | | ORNITHINE DECARBOXYLASE FROM TRYPANOSOMA BRUCEI K69A MUTANT IN COMPLEX WITH ALPHA-DIFLUOROMETHYLORNITHINE | | Descriptor: | ALPHA-DIFLUOROMETHYLORNITHINE, PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Grishin, N.V, Osterman, A.L, Brooks, H.B, Phillips, M.A, Goldsmith, E.J. | | Deposit date: | 1999-05-18 | | Release date: | 1999-11-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of ornithine decarboxylase from Trypanosoma brucei: the native structure and the structure in complex with alpha-difluoromethylornithine.
Biochemistry, 38, 1999
|
|
1CMP
 
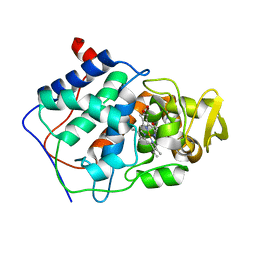 | | SMALL MOLECULE BINDING TO AN ARTIFICIALLY CREATED CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE | | Descriptor: | 2,3-DIMETHYLIMIDAZOLIUM ION, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Mcree, D.E, Churchill, M.J, Goodin, D.B. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small molecule binding to an artificially created cavity at the active site of cytochrome c peroxidase.
Biochemistry, 33, 1994
|
|
3N29
 
 | | Crystal structure of carboxynorspermidine decarboxylase complexed with Norspermidine from Campylobacter jejuni | | Descriptor: | Carboxynorspermidine decarboxylase, GLYCEROL, N-(3-aminopropyl)propane-1,3-diamine, ... | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
2CNF
 
 | | Structural Insights into the Design of Nonpeptidic Isothiazolidinone- Containing Inhibitors of Protein Tyrosine Phosphatase 1B | | Descriptor: | (5S)-5-{4-[(2S)-2-(1H-BENZIMIDAZOL-2-YL)-2-(1,3-BENZOTHIAZOL-2-YLAMINO)ETHYL]PHENYL}ISOTHIAZOLIDIN-3-ONE 1,1-DIOXIDE, MAGNESIUM ION, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Ala, P.J, Gonneville, L, Hillman, M, Becker-Pasha, M, Yue, E.W, Douty, B, Wayland, B, Polam, P, Crawley, M.L, McLaughlin, E, Sparks, R.B, Glass, B, Takvorian, A, Combs, A.P, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-21 | | Release date: | 2006-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into the Design of Nonpeptidic Isothiazolidinone-Containing Inhibitors of Protein- Tyrosine Phosphatase 1B.
J.Biol.Chem., 281, 2006
|
|
2A1B
 
 | | Carboxysome shell protein ccmK2 | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmK homolog 2 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-20 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
2A10
 
 | | carboxysome shell protein ccmK4 | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmK homolog 4 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Tanaka, S, Nguyen, C.V, Phillips, M, Beeby, M, Yeates, T.O. | | Deposit date: | 2005-06-17 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Protein structures forming the shell of primitive bacterial organelles
Science, 309, 2005
|
|
2H3K
 
 | | Solution Structure of the first NEAT domain of IsdH | | Descriptor: | Haptoglobin-binding surface anchored protein | | Authors: | Pilpa, R.M, Fadeev, E.A, Villareal, V.A, Wong, M.A, Phillips, M, Clubb, R.T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the NEAT (NEAr Transporter) domain from IsdH/HarA: the human hemoglobin receptor in Staphylococcus aureus.
J.Mol.Biol., 360, 2006
|
|
2HUE
 
 | | Structure of the H3-H4 chaperone Asf1 bound to histones H3 and H4 | | Descriptor: | Anti-silencing protein 1, GLYCEROL, Histone H3, ... | | Authors: | English, C.M, Churchill, M.E.A, Tyler, J.K. | | Deposit date: | 2006-07-26 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the histone chaperone activity of asf1.
Cell(Cambridge,Mass.), 127, 2006
|
|
1NW6
 
 | | Structure of the beta class N6-adenine DNA methyltransferase RsrI bound to sinefungin | | Descriptor: | CHLORIDE ION, MODIFICATION METHYLASE RSRI, SINEFUNGIN | | Authors: | Thomas, C.B, Scavetta, R.D, Gumport, R.I, Churchill, M.E.A. | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding
J.Biol.Chem., 278, 2003
|
|
1NW8
 
 | | Structure of L72P mutant beta class N6-adenine DNA methyltransferase RsrI | | Descriptor: | CHLORIDE ION, MODIFICATION METHYLASE RSRI | | Authors: | Thomas, C.B, Scavetta, R.D, Gumport, R.I, Churchill, M.E.A. | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding
J.Biol.Chem., 278, 2003
|
|
2NV9
 
 | | The X-ray Crystal Structure of the Paramecium bursaria Chlorella virus arginine decarboxylase | | Descriptor: | A207R protein, arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shah, R.H, Akella, R, Goldsmith, E, Phillips, M.A. | | Deposit date: | 2006-11-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray Structure of Paramecium bursaria Chlorella Virus Arginine Decarboxylase: Insight into the Structural Basis for Substrate Specificity.
Biochemistry, 46, 2007
|
|
1QRV
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF HMG-D AND DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*AP*TP*CP*GP*C)-3'), HIGH MOBILITY GROUP PROTEIN D, SODIUM ION | | Authors: | Murphy IV, F.V, Sweet, R.M, Churchill, M.E.A. | | Deposit date: | 1999-06-15 | | Release date: | 1999-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition.
EMBO J., 18, 1999
|
|
3FGH
 
 | | Human mitochondrial transcription factor A box B | | Descriptor: | CADMIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Gangelhoff, T.A, Mungalachetty, P, Nix, J, Churchill, M.E.A. | | Deposit date: | 2008-12-06 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis and DNA binding of the HMG domains of the human mitochondrial transcription factor A
Nucleic Acids Res., 37, 2009
|
|
1F3T
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA BRUCEI ORNITHINE DECARBOXYLASE (ODC) COMPLEXED WITH PUTRESCINE, ODC'S REACTION PRODUCT. | | Descriptor: | 1,4-DIAMINOBUTANE, ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jackson, L.K, Brooks, H.B, Osterman, A.L, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2000-06-06 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Altering the reaction specificity of eukaryotic ornithine decarboxylase.
Biochemistry, 39, 2000
|
|
3I6R
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase bound with triazolopyrimidine-based inhibitor DSM74 | | Descriptor: | 5-methyl-N-[4-(trifluoromethyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase homolog, mitochondrial, ... | | Authors: | Deng, X, Phillips, M.A. | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural plasticity of malaria dihydroorotate dehydrogenase allows selective binding of diverse chemical scaffolds.
J.Biol.Chem., 284, 2009
|
|
3I68
 
 | |
3I65
 
 | | Plasmodium falciparum dihydroorotate dehydrogenase bound with triazolopyrimidine-based inhibitor DSM1 | | Descriptor: | 5-methyl-7-(naphthalen-2-ylamino)-1H-[1,2,4]triazolo[1,5-a]pyrimidine-3,8-diium, Dihydroorotate dehydrogenase homolog, mitochondrial, ... | | Authors: | Deng, X, Phillips, M.A. | | Deposit date: | 2009-07-06 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural plasticity of malaria dihydroorotate dehydrogenase allows selective binding of diverse chemical scaffolds.
J.Biol.Chem., 284, 2009
|
|
3MAZ
 
 | | Crystal Structure of the Human BRDG1/STAP-1 SH2 Domain in Complex with the NTAL pTyr136 Peptide | | Descriptor: | CheD family protein, MALONATE ION, Signal-transducing adaptor protein 1 | | Authors: | Kaneko, T, Huang, H, Zhao, B, Li, L, Liu, H, Voss, C.K, Wu, C, Schiller, M.R, Li, S.S. | | Deposit date: | 2010-03-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Loops govern SH2 domain specificity by controlling access to binding pockets.
Sci.Signal., 3, 2010
|
|
3PLS
 
 | | RON in complex with ligand AMP-PNP | | Descriptor: | MAGNESIUM ION, Macrophage-stimulating protein receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, J, Steinbacher, S, Augustin, M, Schreiner, P, Epstein, D, Mulvihill, M.J, Crew, A.P. | | Deposit date: | 2010-11-15 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Crystal Structure of a Constitutively Active Mutant RON Kinase Suggests an Intramolecular Autophosphorylation Hypothesis
Biochemistry, 49, 2010
|
|
