2BW3
 
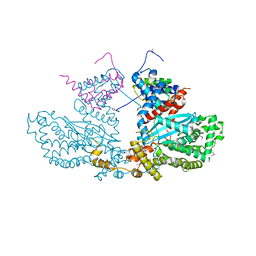 | | Three-dimensional structure of the Hermes DNA transposase | | 分子名称: | TRANSPOSASE | | 著者 | Hickman, A.B, Perez, Z.N, Zhou, L, Musingarimi, P, Ghirlando, R, Hinshaw, J.E, Craig, N.L, Dyda, F. | | 登録日 | 2005-07-11 | | 公開日 | 2005-07-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular Architecture of a Eukaryotic DNA Transposase
Nat.Struct.Mol.Biol., 12, 2005
|
|
1F1Z
 
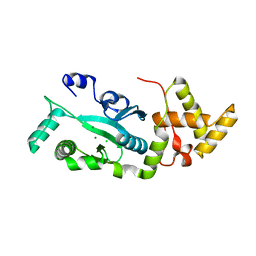 | | TNSA, a catalytic component of the TN7 transposition system | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, TNSA ENDONUCLEASE | | 著者 | Hickman, A.B, Li, Y, Mathew, S.V, May, E.W, Craig, N.L, Dyda, F. | | 登録日 | 2000-05-21 | | 公開日 | 2000-06-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Unexpected structural diversity in DNA recombination: the restriction endonuclease connection.
Mol.Cell, 5, 2000
|
|
1CJW
 
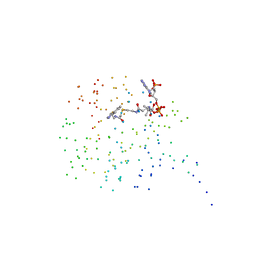 | | SEROTONIN N-ACETYLTRANSFERASE COMPLEXED WITH A BISUBSTRATE ANALOG | | 分子名称: | COA-S-ACETYL TRYPTAMINE, PROTEIN (SEROTONIN N-ACETYLTRANSFERASE) | | 著者 | Hickman, A.B, Namboodiri, M.A.A, Klein, D.C, Dyda, F. | | 登録日 | 1999-04-19 | | 公開日 | 1999-05-06 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structural basis of ordered substrate binding by serotonin N-acetyltransferase: enzyme complex at 1.8 A resolution with a bisubstrate analog.
Cell(Cambridge,Mass.), 97, 1999
|
|
1RZ9
 
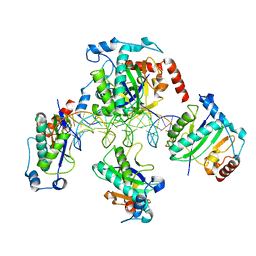 | | Crystal Structure of AAV Rep complexed with the Rep-binding sequence | | 分子名称: | 26-MER, Rep protein | | 著者 | Hickman, A.B, Ronning, D.R, Perez, Z.N, Kotin, R.M, Dyda, F. | | 登録日 | 2003-12-24 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces.
Mol.Cell, 13, 2004
|
|
1M55
 
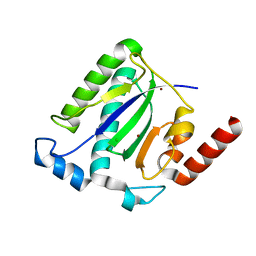 | | Catalytic domain of the Adeno Associated Virus type 5 Rep protein | | 分子名称: | CHLORIDE ION, Rep protein, ZINC ION | | 著者 | Hickman, A.B, Ronning, D.R, Kotin, R.M, Dyda, F. | | 登録日 | 2002-07-08 | | 公開日 | 2002-08-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep.
Mol.Cell, 10, 2002
|
|
2XM3
 
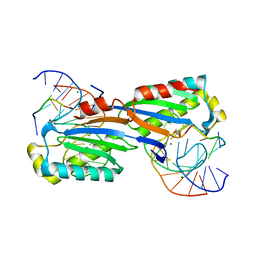 | | Deinococcus radiodurans ISDra2 Transposase Left end DNA complex | | 分子名称: | 5'-D(*TP*TP*AP*GP*T)-3', ACETATE ION, DRA2 TRANSPOSASE BINDING ELEMENT, ... | | 著者 | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | 登録日 | 2010-07-22 | | 公開日 | 2010-10-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2XQC
 
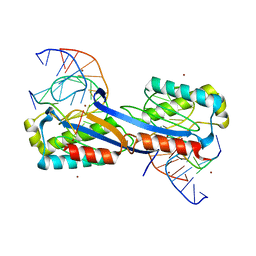 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE COMPLEXED WITH LEFT END RECOGNITION AND CLEAVAGE SITE AND ZN | | 分子名称: | 5'-D(TP*TP*GP*AP*TP*GP)-3', DRA2 TRANSPOSASE LEFT END RECOGNITION SEQUENCE, TRANSPOSASE, ... | | 著者 | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | 登録日 | 2010-09-01 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2XMA
 
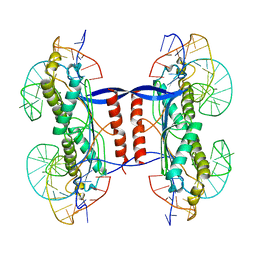 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE RIGHT END DNA COMPLEX | | 分子名称: | DRA2 TRANSPOSASE RIGHT END RECOGNITION SITE, MAGNESIUM ION, TRANSPOSASE | | 著者 | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | 登録日 | 2010-07-26 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2XO6
 
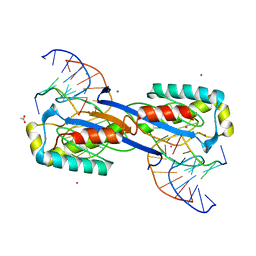 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE Y132F MUTANT COMPLEXED WITH LEFT END RECOGNITION AND CLEAVAGE SITE | | 分子名称: | 5'-D(*TP*TP*GP*AP*TP*G)-3', ACETATE ION, CADMIUM ION, ... | | 著者 | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | 登録日 | 2010-08-09 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
1AIH
 
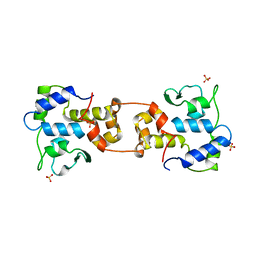 | | CATALYTIC DOMAIN OF BACTERIOPHAGE HP1 INTEGRASE | | 分子名称: | HP1 INTEGRASE, MAGNESIUM ION, SULFATE ION | | 著者 | Hickman, A.B, Waninger, S, Scocca, J.J, Dyda, F. | | 登録日 | 1997-04-17 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 A resolution.
Cell(Cambridge,Mass.), 89, 1997
|
|
1B6B
 
 | |
4D1Q
 
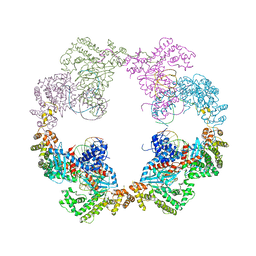 | | Hermes transposase bound to its terminal inverted repeat | | 分子名称: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | 著者 | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | 登録日 | 2014-05-04 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
7TH0
 
 | |
5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | 分子名称: | ZINC ION, piggyBac transposase | | 著者 | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | 登録日 | 2016-07-30 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
1T0F
 
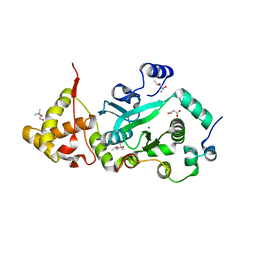 | | Crystal Structure of the TnsA/TnsC(504-555) complex | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, MALONIC ACID, ... | | 著者 | Ronning, D.R, Li, Y, Perez, Z.N, Ross, P.D, Hickman, A.B, Craig, N.L, Dyda, F. | | 登録日 | 2004-04-08 | | 公開日 | 2004-11-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The carboxy-terminal portion of TnsC activates the Tn7 transposase through a specific interaction with TnsA.
Embo J., 23, 2004
|
|
4ER8
 
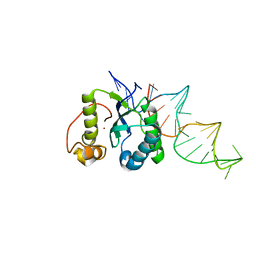 | | Structure of the REP associates tyrosine transposase bound to a REP hairpin | | 分子名称: | DNA (32-MER), NICKEL (II) ION, TnpArep for protein | | 著者 | Messing, S.A.J, Ton-Hoang, B, Hickman, A.B, Ghirlando, R, Chandler, M, Dyda, F. | | 登録日 | 2012-04-19 | | 公開日 | 2012-08-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The processing of repetitive extragenic palindromes: the structure of a repetitive extragenic palindrome bound to its associated nuclease.
Nucleic Acids Res., 40, 2012
|
|
1ITG
 
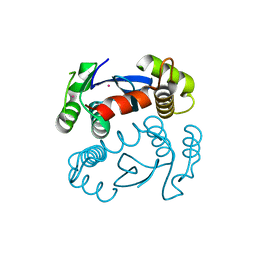 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HIV-1 INTEGRASE: SIMILARITY TO OTHER POLYNUCLEOTIDYL TRANSFERASES | | 分子名称: | CACODYLATE ION, HIV-1 INTEGRASE | | 著者 | Dyda, F, Hickman, A.B, Jenkins, T.M, Engelman, A, Craigie, R, Davies, D.R. | | 登録日 | 1994-11-21 | | 公開日 | 1995-05-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases.
Science, 266, 1994
|
|
1UUT
 
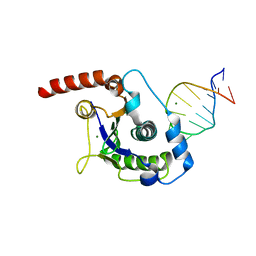 | | The Nuclease Domain of Adeno-Associated Virus Rep Complexed with the RBE' Stemloop of the Viral Inverted Terminal Repeat | | 分子名称: | 5'-D(*CP*AP*GP*CP*TP*CP*TP*TP*TP*GP *AP*GP*CP*TP*G)-3', CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Dyda, F, Hickman, A.B, Ronning, D.R, Perez, Z.N, Kotin, R.M. | | 登録日 | 2004-01-10 | | 公開日 | 2004-02-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Nuclease Domain of Adeno-Associated Virus Rep Coordinates Replication Initiation Using Two Distinct DNA Recognition Interfaces
Mol.Cell, 13, 2004
|
|
1BIS
 
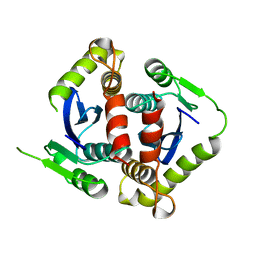 | | HIV-1 INTEGRASE CORE DOMAIN | | 分子名称: | HIV-1 INTEGRASE | | 著者 | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | 登録日 | 1998-06-19 | | 公開日 | 1998-08-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIZ
 
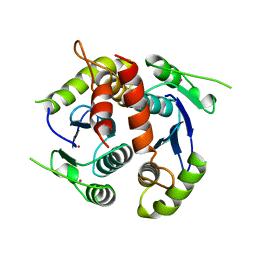 | | HIV-1 INTEGRASE CORE DOMAIN | | 分子名称: | CACODYLATE ION, HIV-1 INTEGRASE | | 著者 | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | 登録日 | 1998-06-21 | | 公開日 | 1998-08-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIU
 
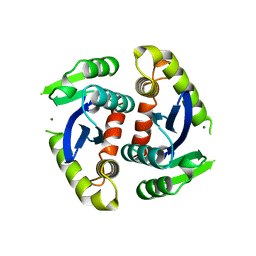 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | 分子名称: | HIV-1 INTEGRASE, MAGNESIUM ION | | 著者 | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | 登録日 | 1998-06-19 | | 公開日 | 1998-08-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6X67
 
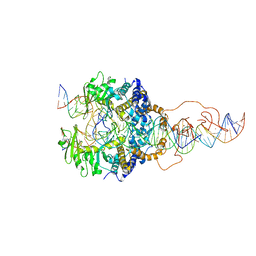 | | Cryo-EM structure of piggyBac transposase strand transfer complex (STC) | | 分子名称: | CALCIUM ION, DNA (37-MER), DNA (47-MER), ... | | 著者 | Chen, Q, Hickman, A.B, Dyda, F. | | 登録日 | 2020-05-27 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Structural basis of seamless excision and specific targeting by piggyBac transposase
Nat Commun, 11, 2020
|
|
6X68
 
 | | Cryo-EM structure of piggyBac transposase synaptic complex with hairpin DNA (SNHP) | | 分子名称: | CALCIUM ION, Transposase, ZINC ION, ... | | 著者 | Chen, Q, Hickman, A.B, Dyda, F. | | 登録日 | 2020-05-27 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Structural basis of seamless excision and specific targeting by piggyBac transposase
Nat Commun, 11, 2020
|
|
6OPM
 
 | | Casposase bound to integration product | | 分子名称: | CALCIUM ION, CRISPR-associated endonuclease Cas1, DNA 21-mer, ... | | 著者 | Dyda, F, Hickman, A.B, Kailasan, S. | | 登録日 | 2019-04-25 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-08-26 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Casposase structure and the mechanistic link between DNA transposition and spacer acquisition by CRISPR-Cas.
Elife, 9, 2020
|
|
6DWZ
 
 | | Hermes transposase deletion dimer complex with (C/G) DNA | | 分子名称: | DNA (26-MER), DNA (5'-D(*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*C)-3'), ... | | 著者 | Dyda, F, Hickman, A.B. | | 登録日 | 2018-06-28 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
