1QK5
 
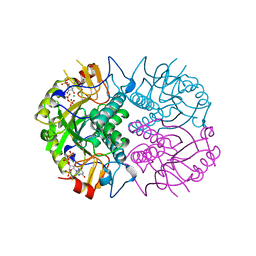 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE WITH XMP, PYROPHOSPHATE AND TWO MG2+ IONS | | Descriptor: | HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Heroux, A, White, E.L, Ross, L.J, Davis, R.L, Borhani, D.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Toxoplasma Gondii Hypoxanthine-Guanine Phosphoribosyltransferase with Xmp, Pyrophosphate and Two Mg2+ Ions Bound: Insights Into the Catalytic Mechanism
Biochemistry, 38, 1999
|
|
1QK3
 
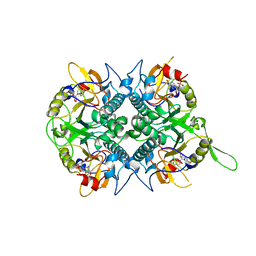 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE GMP COMPLEX | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Heroux, A, White, E.L, Ross, L.J, Borhani, D.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the Toxoplasma Gondii Hypoxanthine-Guanine Phosphoribosyltransferase Gmp and -Imp Complexes: Comparison of Purine Binding Interactions with the Xmp Complex
Biochemistry, 38, 1999
|
|
1FSG
 
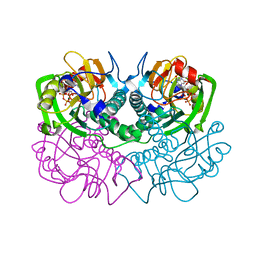 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE COMPLEXED WITH 9-DEAZAGUANINE, ALPHA-D-5-PHOSPHORIBOSYL-1-PYROPHOSPHATE (PRPP) AND TWO MG2+ IONS | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAGUANINE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, ... | | Authors: | Heroux, A, White, E.L, Ross, L.J, Kuzin, A.P, Borhani, D.W. | | Deposit date: | 2000-09-08 | | Release date: | 2000-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Substrate deformation in a hypoxanthine-guanine phosphoribosyltransferase ternary complex: the structural basis for catalysis.
Structure Fold.Des., 8, 2000
|
|
1QK4
 
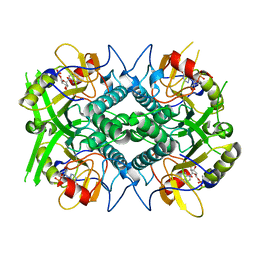 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE IMP COMPLEX | | Descriptor: | HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, INOSINIC ACID | | Authors: | Heroux, A, White, E.L, Ross, L.J, Borhani, D.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of the Toxoplasma Gondii Hypoxanthine-Guanine Phosphoribosyltransferase Gmp and -Imp Complexes: Comparison of Purine Binding Interactions with the Xmp Complex
Biochemistry, 38, 1999
|
|
3D9E
 
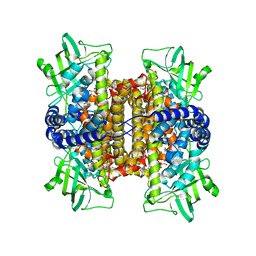 | | Nitroalkane oxidase: active site mutant D402N crystallized with 1-nitrooctane | | Descriptor: | 1-nitrooctane, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
3D9D
 
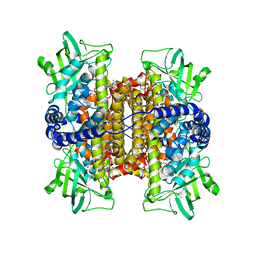 | | Nitroalkane oxidase: mutant D402N crystallized with 1-nitrohexane | | Descriptor: | 1-nitrohexane, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
3D9F
 
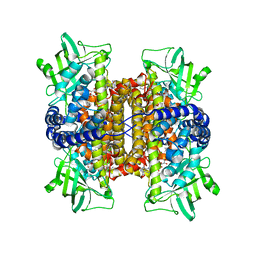 | | Nitroalkane oxidase: active site mutant S276A crystallized with 1-nitrohexane | | Descriptor: | 1-nitrohexane, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
3D9G
 
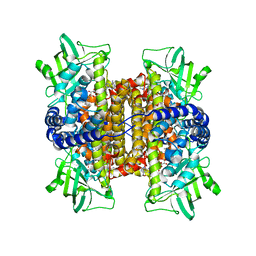 | | Nitroalkane oxidase: wild type crystallized in a trapped state forming a cyanoadduct with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Nitroalkane oxidase, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
2R0Y
 
 | | Structure of the Rsc4 tandem bromodomain in complex with an acetylated H3 peptide | | Descriptor: | Chromatin structure-remodeling complex protein RSC4, Histone H3 peptide | | Authors: | VanDemark, A.P, Kasten, M.M, Ferris, E, Heroux, A, Hill, C.P, Cairns, B.R. | | Deposit date: | 2007-08-21 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation.
Mol.Cell, 27, 2007
|
|
7Q93
 
 | | Crystal Structure of Agrobacterium tumefaciens NADQ, NAD complex. | | Descriptor: | GLYCEROL, NADQ transcription factor, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
7Q92
 
 | | Crystal Structure of Agrobacterium tumefaciens NADQ, ATP complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NADQ transcription factor, POTASSIUM ION, ... | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
1R7S
 
 | | PUTIDAREDOXIN (Fe2S2 ferredoxin), C73G mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Smith, N, Mayhew, M, Kelly, H, Robinson, H, Heroux, A, Holden, M.J, Gallagher, D.T. | | Deposit date: | 2003-10-22 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of C73G putidaredoxin from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4XGK
 
 | | Crystal structure of UDP-galactopyranose mutase from Corynebacterium diphtheriae in complex with 2-[4-(4-chlorophenyl)-7-(2-thienyl)-2-thia-5,6,8,9-tetrazabicyclo[4.3.0]nona-4,7,9-trien-3-yl]acetic | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, SULFATE ION, UDP-galactopyranose mutase, ... | | Authors: | Wangkanont, K, Heroux, A, Forest, K.T, Kiessling, L.L. | | Deposit date: | 2014-12-31 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Virtual Screening for UDP-Galactopyranose Mutase Ligands Identifies a New Class of Antimycobacterial Agents.
Acs Chem.Biol., 10, 2015
|
|
8ASH
 
 | | Crystal structure of d(CCGGGGTACCCCGG) with XRB | | Descriptor: | 4-[(~{E})-(3,6-dimethyl-1,3-benzothiazol-2-yl)iminomethyl]-~{N},~{N}-dimethyl-aniline, DNA (5'-D(*CP*CP*GP*GP*GP*GP*TP*AP*CP*CP*CP*CP*GP*G)-3') | | Authors: | Sbirkova-Dimitrova, H.I, Shivachev, B.L, Rusev, R, Kuvandjiev, N, Heroux, A, Doukov, T. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.837 Å) | | Cite: | Structural Characterization of Alzheimer DNA Promoter Sequences from the Amyloid Precursor Gene in the Presence of Thioflavin T and Analogs
Crystals, 12, 2022
|
|
8ASK
 
 | | Crystal structure of d(GCCCACCACGGC) | | Descriptor: | DNA (5'-D(P*GP*CP*CP*CP*AP*CP*CP*AP*CP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*GP*TP*GP*GP*TP*GP*GP*GP*C)-3') | | Authors: | Sbirkova-Dimitrova, H.I, Shivachev, B.L, Rusev, R, Heroux, A, Doukov, T, Kuvandjiev, N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Structural Characterization of Alzheimer DNA Promoter Sequences from the Amyloid Precursor Gene in the Presence of Thioflavin T and Analogs
Crystals, 12, 2022
|
|
7Q94
 
 | | Crystal Structure of Agrobacterium tumefaciens NADQ, DNA complex. | | Descriptor: | DNA binding region (31-MER), NADQ transcription factor | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
7Q91
 
 | | Crystal Structure of Agrobacterium tumefaciens NADQ, native form. | | Descriptor: | NADQ transcription factor, SODIUM ION | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
5JOB
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25K/I92A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Caro, J.A, Heroux, A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2016-05-02 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25K/I92A at cryogenic temperature
To be Published
|
|
5KEE
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25K/I92F at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Caro, J.A, Heroux, A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25K/I92F at cryogenic temperature
To be Published
|
|
4R0X
 
 | | Allosteric coupling of conformational transitions in the FK1 domain of FKBP51 near the site of steroid receptor interaction | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | LeMaster, D.M, Mustafi, S.M, Brecher, M, Zhang, J, Heroux, A, Li, H.M, Hernandez, G. | | Deposit date: | 2014-08-02 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Coupling of Conformational Transitions in the N-terminal Domain of the 51-kDa FK506-binding Protein (FKBP51) Near Its Site of Interaction with the Steroid Receptor Proteins.
J.Biol.Chem., 290, 2015
|
|
8P5K
 
 | | Kinase domain of mutant human ULK1 in complex with compound MRT68921 | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Battista, T, Semrau, M.S, Heroux, A, Lolli, G, Storici, P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Crystal structures of ULK1 in complex with KCGS compounds
To Be Published
|
|
5HUR
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25T/I92K at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Caro, J.A, Heroux, A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2016-01-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25T/I92K at cryogenic temperature
To be Published
|
|
2P8R
 
 | | Crystal structure of the C-terminal domain of C. elegans pre-mRNA splicing factor Prp8 carrying R2303K mutant | | Descriptor: | Pre-mRNA-splicing factor Prp8 | | Authors: | Zhang, L, Shen, J, Guarnieri, M.T, Heroux, A, Yang, K, Zhao, R. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants
Protein Sci., 16, 2007
|
|
2P87
 
 | | Crystal structure of the C-terminal domain of C. elegans pre-mRNA splicing factor Prp8 | | Descriptor: | Pre-mRNA-splicing factor Prp8 | | Authors: | Zhang, L, Shen, J, Guarnieri, M.T, Heroux, A, Yang, K, Zhao, R. | | Deposit date: | 2007-03-21 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants
Protein Sci., 16, 2007
|
|
1KMV
 
 | | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND (Z)-6-(2-[2,5-DIMETHOXYPHENYL]ETHEN-1-YL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE (SRI-9662), A LIPOPHILIC ANTIFOLATE | | Descriptor: | (Z)-6-(2-[2,5-DIMETHOXYPHENYL]ETHEN-1-YL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, DIMETHYL SULFOXIDE, ... | | Authors: | Klon, A.E, Heroux, A, Ross, L.J, Pathak, V, Johnson, C.A, Piper, J.R, Borhani, D.W. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic structures of human dihydrofolate reductase complexed with NADPH and two lipophilic antifolates at 1.09 a and 1.05 a resolution.
J.Mol.Biol., 320, 2002
|
|
