2YP6
 
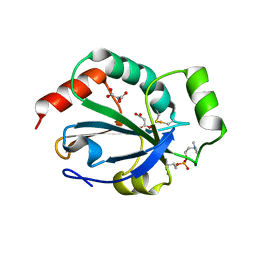 | | Crystal structure of the pneumoccocal exposed lipoprotein thioredoxin sp_1000 (Etrx2) from Streptococcus pneumoniae strain TIGR4 in complex with Cyclofos 3 TM | | Descriptor: | 3-Cyclohexyl-1-Propylphosphocholine, MALONATE ION, THIOREDOXIN FAMILY PROTEIN | | Authors: | Bartual, S.G, Shalek, M, Abdullah, M.R, Jensch, I, Asmat, T.M, Petruschka, L, Pribyl, T, Hermoso, J.A, Hammerschmidt, S. | | Deposit date: | 2012-10-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.767 Å) | | Cite: | Molecular Architecture of Streptococcus Pneumoniae Surface Thioredoxin-Fold Lipoproteins Crucial for Extracellular Oxidative Stress Resistance and Maintenance of Virulence.
Embo Mol.Med., 5, 2013
|
|
5OAU
 
 | |
5OJ0
 
 | |
7AGZ
 
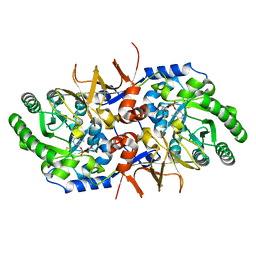 | | BsrV no-histagged | | Descriptor: | Broad specificity amino-acid racemase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Carrasco-Lopez, C, Rojas-Altuve, A, Espaillat, A, Cava, F, Hermoso, J.A. | | Deposit date: | 2020-09-23 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Binding of non-canonical peptidoglycan controls Vibrio cholerae broad spectrum racemase activity.
Comput Struct Biotechnol J, 19, 2021
|
|
5AA4
 
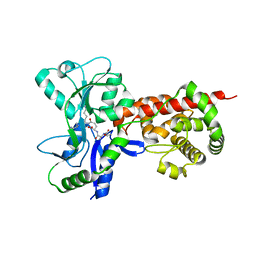 | | Crystal structure of MltF from Pseudomonas aeruginosa in complex with cell-wall tetrapeptide | | Descriptor: | MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE F, [6-[[(2~{R})-1-azanyl-1-oxidanylidene-propan-2-yl]amino]-6-oxidanylidene-5-[[(4~{R})-5-oxidanyl-5-oxidanylidene-4-[[(2~{S})-2-[[(2~{R})-2-oxidanylpropanoyl]amino]propanoyl]amino]pentanoyl]amino]hexyl]azanium | | Authors: | Dominguez-Gil, T, Acebron, I, Hermoso, J.A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation by Allostery in Cell-Wall Remodeling by a Modular Membrane-Bound Lytic Transglycosylase from Pseudomonas aeruginosa.
Structure, 24, 2016
|
|
3ZHG
 
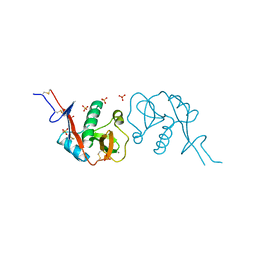 | |
5CYB
 
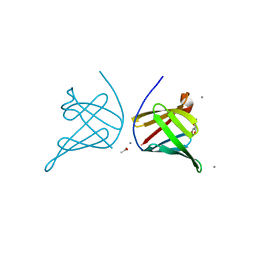 | | Structure of a lipocalin lipoprotein affecting virulence in Streptococcus pneumoniae | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Carrasco-Lopez, C, Abdullah, M.R, Hammerschmidt, S, Hermoso, J.A. | | Deposit date: | 2015-07-30 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unraveling the function and structure of a lipocalin lipoprotein affecting virulence in Streptococcus pneumoniae
To Be Published
|
|
6GI4
 
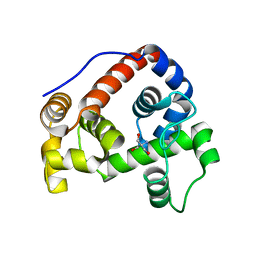 | |
2W5U
 
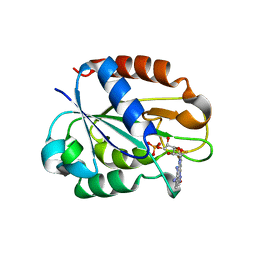 | | Flavodoxin from Helicobacter pylori in complex with the C3 inhibitor | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, [2-(5-amino-4-cyano-1H-pyrazol-1-yl)-5-(trifluoromethyl)phenyl](hydroxy)oxoammonium | | Authors: | Cremades, N, Perez-Dorado, I, Hermoso, J.A, Martinez-Julvez, M, Sancho, J. | | Deposit date: | 2008-12-12 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of Specific Flavodoxin Inhibitors as Potential Therapeutic Agents Against Helicobacter Pylori Infection.
Acs Chem.Biol., 4, 2009
|
|
1QM4
 
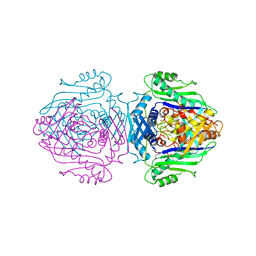 | | Methionine Adenosyltransferase Complexed with a L-Methionine Analogue | | Descriptor: | L-2-AMINO-4-METHOXY-CIS-BUT-3-ENOIC ACID, MAGNESIUM ION, METHIONINE ADENOSYLTRANSFERASE, ... | | Authors: | Gonzalez, B, Pajares, M.A, Hermoso, J.A, Sanz-Aparicio, J. | | Deposit date: | 1999-09-20 | | Release date: | 2000-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The Crystal Structure of Tetrameric Methionine Adenosyltransferase from Rat Liver Reveals the Methionine-Binding Site
J.Mol.Biol., 300, 2000
|
|
2Y8P
 
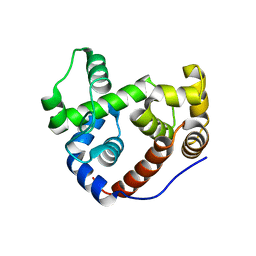 | | Crystal Structure of an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase (MltE) from Escherichia coli | | Descriptor: | ENDO-TYPE MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE A | | Authors: | Artola-Recolons, C, Carrasco-Lopez, C, Llarrull, L.I, Kumarasiri, M, Lastochkin, E, Martinez-Ilarduya, I, Meindl, K, Uson, I, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2011-02-08 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | High-Resolution Crystal Structure of Mlte, an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase from Escherichia Coli.
Biochemistry, 50, 2011
|
|
5A89
 
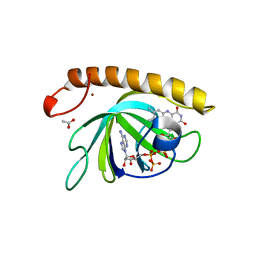 | | Crystal structure of the riboflavin kinase module of FAD synthetase from Corynebacterium ammoniagenes in complex with FMN and ADP(P 21 21 21) | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Medina, M. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights Into the Synthesis of Fmn in Prokaryotic Organisms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1O71
 
 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II complexed with glycerol | | Descriptor: | GLYCEROL, STICHOLYSIN II | | Authors: | Mancheno, J.M, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-10-23 | | Release date: | 2003-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
2WC1
 
 | | Three-dimensional Structure of the Nitrogen Fixation Flavodoxin (NifF) from Rhodobacter capsulatus at 2.2 A | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Perez-Dorado, I, Bittel, C, Hermoso, J.A, Cortez, N, Carrillo, N. | | Deposit date: | 2009-03-06 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and Phylogenetic Analysis of Rhodobacter Capsulatus Niff: Uncovering General Features of Nitrogen-Fixation (Nif)-Flavodoxins.
Int.J.Mol.Sci., 14, 2013
|
|
1O72
 
 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II complexed with phosphorylcholine | | Descriptor: | PHOSPHOCHOLINE, STICHOLYSIN II | | Authors: | Mancheno, J.M, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-10-23 | | Release date: | 2003-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
5A8A
 
 | | Crystal structure of the riboflavin kinase module of FAD synthetase from Corynebacterium ammoniagenes in complex with FMN and ADP (P3 2 21) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Medina, M. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Synthesis of Fmn in Prokaryotic Organisms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7BAY
 
 | |
5A88
 
 | | Crystal structure of the riboflavin kinase module of FAD synthetase from Corynebacterium ammoniagenes in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Medina, M. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Insights Into the Synthesis of Fmn in Prokaryotic Organisms.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6FCU
 
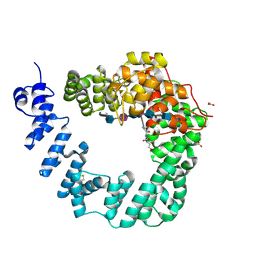 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with 4(NAG-NAMpentapeptide) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ALANINE, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FC4
 
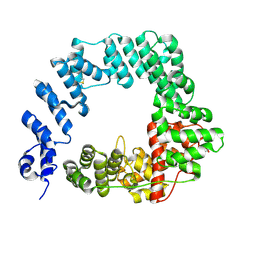 | |
6FCQ
 
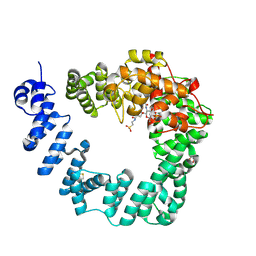 | |
2VNK
 
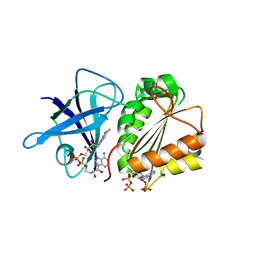 | |
6FCS
 
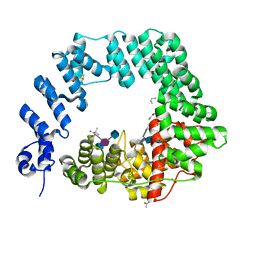 | | The X-ray Structure of Lytic Transglycosylase Slt inactive mutant E503Q from Pseudomonas aeruginosa in complex with NAG-NAMpentapeptide-NAG-NAMpentapeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-3-O-[(2R)-1-amino-1-oxopropan-2-yl]-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ACETATE ION, ... | | Authors: | Batuecas, M.T, Dominguez-Gil, T, Hermoso, J.A. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Exolytic and endolytic turnover of peptidoglycan by lytic transglycosylase Slt ofPseudomonas aeruginosa.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1GZ7
 
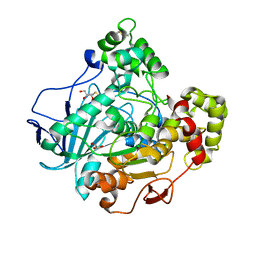 | | Crystal structure of the closed state of lipase 2 from Candida rugosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LIPASE 2 | | Authors: | Mancheno, J.M, Hermoso, J.A. | | Deposit date: | 2002-05-17 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights Into the Lipase/Esterase Behavior in the Candida Rugosa Lipases Family: Crystal Structure of the Lipase 2 Isoenzyme at 1.97A Resolution
J.Mol.Biol., 332, 2003
|
|
1GWY
 
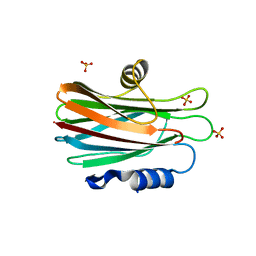 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II | | Descriptor: | STICHOLYSIN II, SULFATE ION | | Authors: | Mancheno, J.M, Martin-Benito, J, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-03-26 | | Release date: | 2003-06-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
