7PL5
 
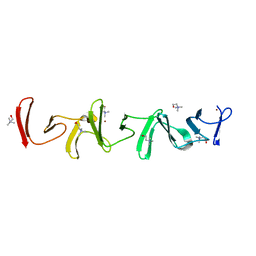 | | Crystal structure of choline-binding module (R1-R9) of LytB from Streptococcus pneumoniae | | Descriptor: | CHOLINE ION, Putative endo-beta-N-acetylglucosaminidase, TRIETHYLENE GLYCOL, ... | | Authors: | Molina, R, Martinez Caballero, S, Hermoso, J.A. | | Deposit date: | 2021-08-28 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis of the final step of cell division in Streptococcus pneumoniae.
Cell Rep, 42, 2023
|
|
5M1A
 
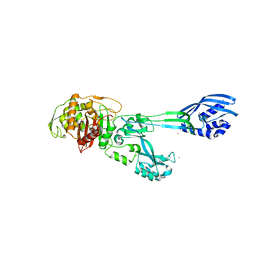 | | Crystal structure of PBP2a from MRSA in the presence of Ceftazidime ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
6YA4
 
 | |
6YAB
 
 | |
6YA3
 
 | |
6Y9U
 
 | |
6YAG
 
 | |
2WC1
 
 | | Three-dimensional Structure of the Nitrogen Fixation Flavodoxin (NifF) from Rhodobacter capsulatus at 2.2 A | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Perez-Dorado, I, Bittel, C, Hermoso, J.A, Cortez, N, Carrillo, N. | | Deposit date: | 2009-03-06 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and Phylogenetic Analysis of Rhodobacter Capsulatus Niff: Uncovering General Features of Nitrogen-Fixation (Nif)-Flavodoxins.
Int.J.Mol.Sci., 14, 2013
|
|
2W5U
 
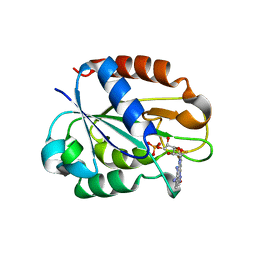 | | Flavodoxin from Helicobacter pylori in complex with the C3 inhibitor | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, [2-(5-amino-4-cyano-1H-pyrazol-1-yl)-5-(trifluoromethyl)phenyl](hydroxy)oxoammonium | | Authors: | Cremades, N, Perez-Dorado, I, Hermoso, J.A, Martinez-Julvez, M, Sancho, J. | | Deposit date: | 2008-12-12 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of Specific Flavodoxin Inhibitors as Potential Therapeutic Agents Against Helicobacter Pylori Infection.
Acs Chem.Biol., 4, 2009
|
|
5M19
 
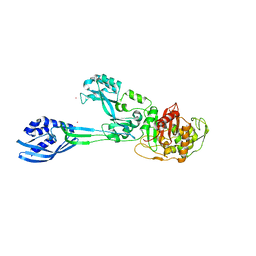 | | Crystal structure of PBP2a from MRSA in the presence of Oxacillin ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
6Z63
 
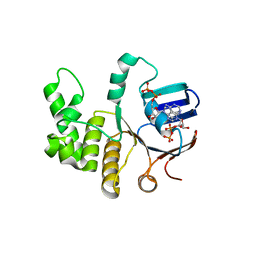 | | FtsE structure from Streptococus pneumoniae in complex with ADP at 1.57 A resolution (spacegroup P 21) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE | | Authors: | Alcorlo, M, Straume, D, Havarstein, L.S, Hermoso, J.A. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae.
Mbio, 11, 2020
|
|
3ZHG
 
 | |
5LY7
 
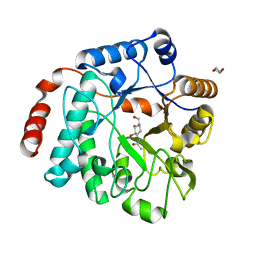 | | Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Beta-hexosaminidase, DI(HYDROXYETHYL)ETHER | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-09-25 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
7PJ4
 
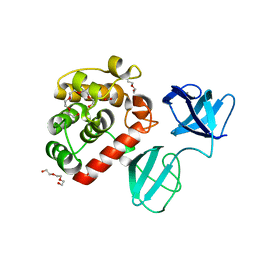 | |
7PJ6
 
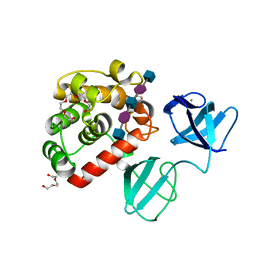 | |
7PJ5
 
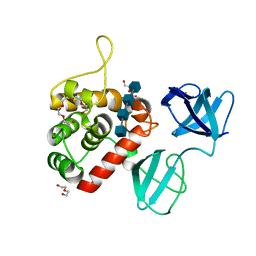 | |
7PJ3
 
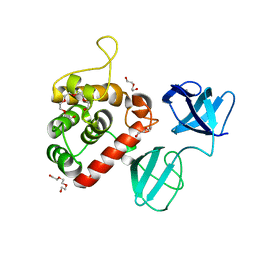 | |
7POD
 
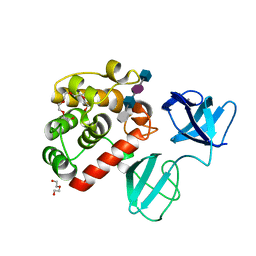 | |
2VNJ
 
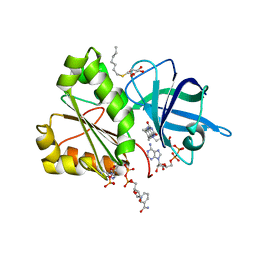 | |
2VNI
 
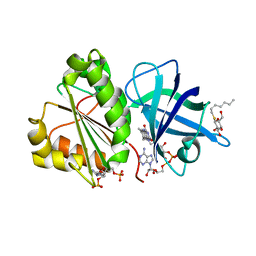 | |
6Z4W
 
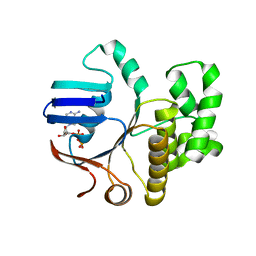 | | FtsE structure from Streptococcus pneumoniae in complex with ADP (space group P 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE | | Authors: | Alcorlo, M, Straume, D, Hermoso, J.A, Havarstein, L.S. | | Deposit date: | 2020-05-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae.
Mbio, 11, 2020
|
|
2VNH
 
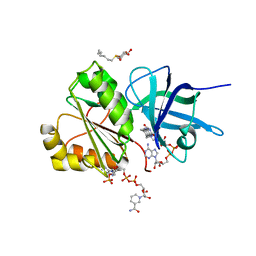 | |
2VNK
 
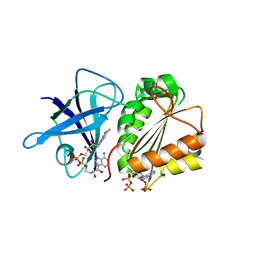 | |
6Z67
 
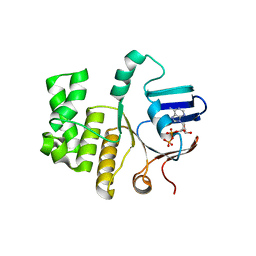 | | FtsE structure of Streptococcus pneumoniae in complex with AMPPNP at 2.4 A resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Alcorlo, M, Straume, D, Havarstein, L.S, Hermoso, j.A. | | Deposit date: | 2020-05-28 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae.
Mbio, 11, 2020
|
|
5G2M
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-HEXOSAMINIDASE | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-04-09 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
