3LLN
 
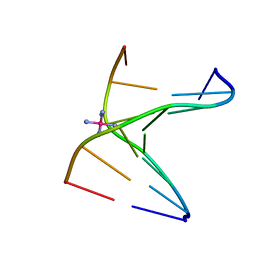 | | Comparison between the orthorhombic an tetragonal form of the heptamer sequence d(GCG(xT)GCG)/d(CGCACGC). | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*CP*GP*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*(XTR)P*GP*CP*G)-3') | | Authors: | Robeyns, K, Herdewijn, P, Van Meervelt, L. | | Deposit date: | 2010-01-29 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comparison between the orthorhombic and tetragonal forms of the heptamer sequence d[GCG(xT)GCG]/d(CGCACGC)
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2BJ6
 
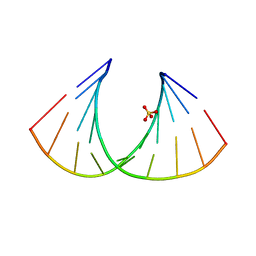 | | Crystal Structure of a decameric HNA-RNA hybrid | | Descriptor: | 5'-R(*GP*GP*CP*AP*UP*UP*AP*CP*GP*GP)-3', SULFATE ION, SYNTHETIC HNA | | Authors: | Maier, T, Przylas, I, Straeter, N, Herdewijn, P, Saenger, W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reinforced Hna Backbone Hydration in the Crystal Structure of a Decameric Hna/RNA Hybrid
J.Am.Chem.Soc., 127, 2005
|
|
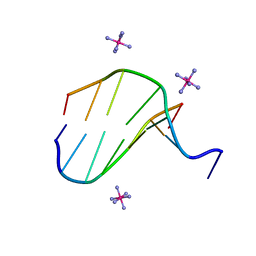
3FL6
 
 | |
1QXB
 
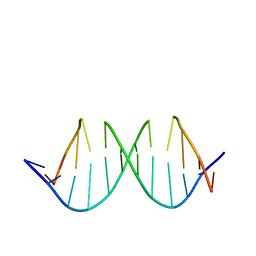 | | NMR structure determination of the self complementary DNA Dodecamer CGCGAATT*CGCG in which a ribose is inserted between the 3'-OH of T8 and the 5'-phosphate group of C9 | | Descriptor: | 5'-d(CpGpCpGpApApTpTpCpGpCpG)-3', beta-D-ribofuranose | | Authors: | Nauwelaerts, K, Vastmans, K, Froeyen, M, Kempeneers, V, Rozenski, J, Rosemeyer, H, Van Aerschot, A, Busson, R, Efimtseva, E, Mikhailov, S, Lescrinier, E, Herdewijn, P. | | Deposit date: | 2003-09-05 | | Release date: | 2004-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cleavage of DNA without loss of genetic information by incorporation of a disaccharide nucleoside.
Nucleic Acids Res., 31, 2003
|
|
1U01
 
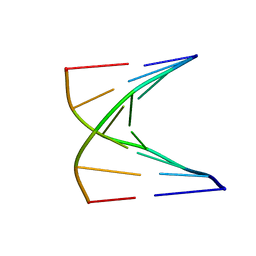 | | High resolution NMR structure of 5-d(GCGT*GCG)-3/5-d(CGCACGC)-3 (T*represents a cyclohexenyl nucleotide) | | Descriptor: | 5'-D(*CP*GP*CP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*(XTR)P*GP*CP*G)-3' | | Authors: | Nauwelaerts, K, Lescrinier, E, Sclep, G, Herdewijn, P. | | Deposit date: | 2004-07-12 | | Release date: | 2005-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Cyclohexenyl nucleic acids: conformationally flexible oligonucleotides.
Nucleic Acids Res., 33, 2005
|
|
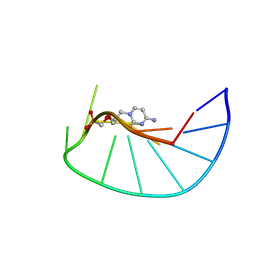
9F3D
 
 | |
1EC4
 
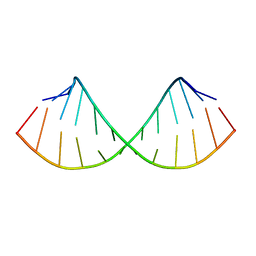 | | SOLUTION STRUCTURE OF A HEXITOL NUCLEIC ACID DUPLEX WITH FOUR CONSECUTIVE T:T BASE PAIRS | | Descriptor: | HEXITOL DODECANUCLEOTIDE | | Authors: | Lescrinier, E, Esnouf, R.M, Schraml, J, Busson, R, Herdewijn, P. | | Deposit date: | 2000-01-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a HNA-RNA hybrid
Chem.Biol., 7, 2000
|
|
1EJZ
 
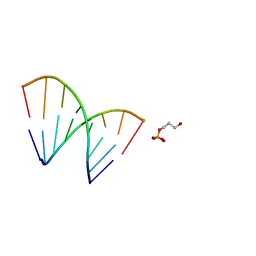 | | SOLUTION STRUCTURE OF A HNA-RNA HYBRID | | Descriptor: | DNA (5'-H(*(6HG)P*(6HC)P*(6HG)P*(6HT)P*(6HA)P*(6HG)P*(6HC)P*(6HG))-3'), PHOSPHORIC ACID MONO-(3-HYDROXY-PROPYL) ESTER, RNA (5'-R(*CP*GP*CP*UP*AP*CP*GP*C)-3') | | Authors: | Lescrnier, E, Esnouf, R, Heus, H.A, Hilbers, C.W, Herdewijn, P. | | Deposit date: | 2000-03-06 | | Release date: | 2000-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a HNA-RNA hybrid.
Chem.Biol., 7, 2000
|
|
1MRN
 
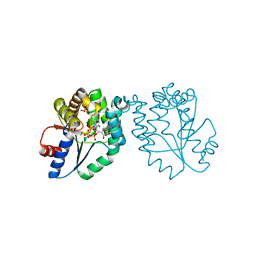 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH BISUBSTRATE INHIBITOR (TP5A) | | Descriptor: | MAGNESIUM ION, P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, SULFATE ION, ... | | Authors: | Haouz, A, Vanheusden, V, Munier-Lehmann, H, Froeyen, M, Herdewijn, P, Van Calenbergh, S, Delarue, M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism.
J.Biol.Chem., 278, 2003
|
|
1KI6
 
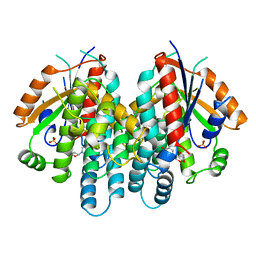 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH A 5-IODOURACIL ANHYDROHEXITOL NUCLEOSIDE | | Descriptor: | 1',5'-ANHYDRO-2',3'-DIDEOXY-2'-(5-IODOURACIL-1-YL)-D-ABABINO-HEXITOL, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Herdewijn, P, Ostrowski, T, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
2KPC
 
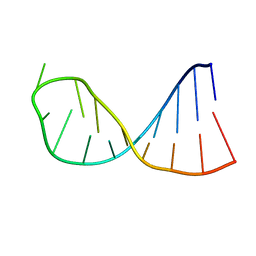 | | Structure determination of the top-loop of the conserved 3 terminal secondary structure in the genome of YFV | | Descriptor: | RNA (5'-R(*UP*GP*AP*GP*CP*AP*CP*AP*GP*UP*UP*UP*GP*CP*UP*CP*A)-3') | | Authors: | Lescrinier, E, Dyubankova, N, Nauwelaerts, K, Jones, R, Herdewijn, P. | | Deposit date: | 2009-10-12 | | Release date: | 2010-06-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of the Top-Loop of the Conserved 3'-Terminal Secondary Structure in the Genome of Flaviviruses.
Chembiochem, 11, 2010
|
|
2MD6
 
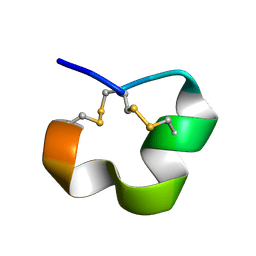 | | NMR SOLUTION STRUCTURE OF ALPHA CONOTOXIN LO1A FROM Conus longurionis | | Descriptor: | ALPHA CONOTOXIN LO1A | | Authors: | Maiti, M, Lescrinier, E, Herdewijn, P, Lebbe, E.K.M, Peigneur, S, D'Souza, L, Tytgat, J. | | Deposit date: | 2013-09-01 | | Release date: | 2014-03-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Function Elucidation of a New alpha-Conotoxin, Lo1a, from Conus longurionis.
J.Biol.Chem., 289, 2014
|
|
2KPD
 
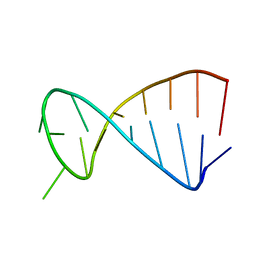 | | Structure determination of the top-loop of the conserved 3 terminal secondary structure in the genome of YFV-mutant | | Descriptor: | RNA (5'-R(*UP*GP*AP*GP*CP*UP*CP*AP*GP*UP*UP*UP*GP*CP*UP*CP*A)-3') | | Authors: | Lescrinier, E, Dyubankova, N, Nauwelaerts, K, Jones, R, Herdewijn, P. | | Deposit date: | 2009-10-12 | | Release date: | 2010-06-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of the Top-Loop of the Conserved 3'-Terminal Secondary Structure in the Genome of Flaviviruses.
Chembiochem, 11, 2010
|
|
2LU6
 
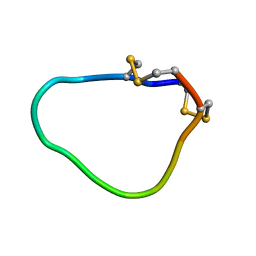 | | NMR solution structure of Midi peptide designed based on m-conotoxins | | Descriptor: | Midi peptide designed based on m-conotoxins | | Authors: | Dyubankova, N, Lescrinier, E, Stevens, M, Tytgat, J, Herdewijn, P, Peigneur, S. | | Deposit date: | 2012-06-08 | | Release date: | 2012-06-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design of bioactive peptides from naturally occurring mu-conotoxin structures.
J.Biol.Chem., 287, 2012
|
|
2MN0
 
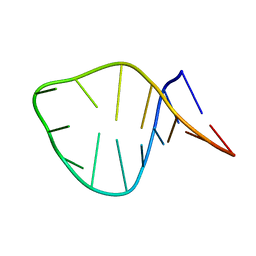 | | D loop of tRNA(Met) | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*(H2U)P*GP*GP*AP*AP*CP*UP*CP*C)-3' | | Authors: | Lescrinier, E, Dyubankova, N, Herdewijn, P. | | Deposit date: | 2014-03-25 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Contribution of dihydrouridine in folding of the D-arm in tRNA.
Org.Biomol.Chem., 13, 2015
|
|
2MSF
 
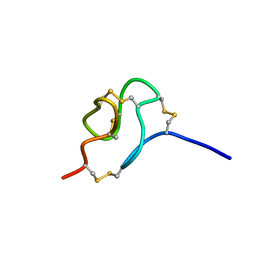 | | NMR SOLUTION STRUCTURE OF SCORPION VENOM TOXIN Ts11 (TsPep1) FROM Tityus serrulatus | | Descriptor: | Peptide TsPep1 | | Authors: | Maiti, M, Lescrinier, E, Herdewijn, P, Cremonez, C.M, Peigneur, S, Cassoli, J.S, Dutra, A.A.A, Waelkens, E, Pimenta, A.M.C, De Lima, M.H, Tytgat, J, Arantes, E.C. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Elucidation of Peptide Ts11 Shows Evidence of a Novel Subfamily of Scorpion Venom Toxins.
Toxins (Basel), 8, 2016
|
|
2N4J
 
 | |
7AII
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND L-METHIONINE TENOFOVIR WITH BOUND MANGANESE | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), Gag-Pol polyprotein, ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
7AHX
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND D-ASPARTATE TENOFOVIR WITH BOUND MANGANESE | | Descriptor: | D-Aspartate Tenofovir, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(*TP*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
7AIG
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND L-GLUTAMATE TENOFOVIR | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), Gag-Pol polyprotein, ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
7AIJ
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND L-METHIONINE TENOFOVIR | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), Gag-Pol polyprotein, ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
7AIF
 
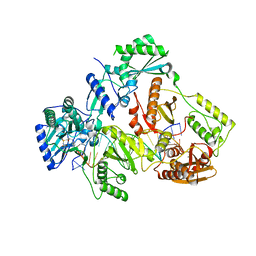 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND L-GLUTAMATE TENOFOVIR WITH BOUND MANGANESE | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), Gag-Pol polyprotein, ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
7AID
 
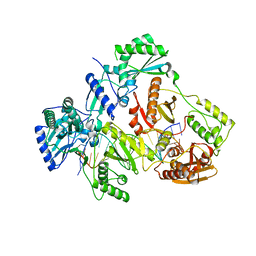 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND D-ASPARTATE TENOFOVIR | | Descriptor: | D-Aspartate Tenofovir, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
7BFX
 
 | | deoxyxylose nucleic acid hairpin | | Descriptor: | dXyNA (5'-D(*(XA)P*(XG)P*(XC)P*(XA)P*(XA)P*(XT)P*(XC)P*(XC)P*(XC)P*(XC)P*(XC)P*(XC)P*(XG)P*(XG)P*(XA)P*(XT)P*(XT)P*(XG)P*(XC)P*T)-3') | | Authors: | Mattelaer, C.-A, Mohitosh, M, Smets, L, Maiti, M, Schepers, G, Mattelaer, H.-P, Rosemeyer, H, Herdewijn, P, Lescrinier, E. | | Deposit date: | 2021-01-05 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | SOLUTION NMR | | Cite: | Stable Hairpin Structures Formed by Xylose-Based Nucleic Acids.
Chembiochem, 22, 2021
|
|
7BFS
 
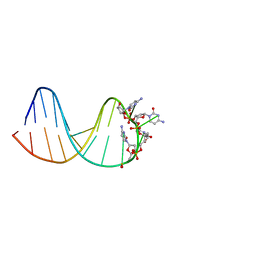 | | deoxyxylose nucleic acid hairpin | | Descriptor: | DNA (5'-D(*AP*GP*CP*AP*AP*TP*CP*CP*(XC)P*(XC)P*(XC)P*(XC)P*GP*GP*AP*TP*TP*GP*CP*T)-3') | | Authors: | Mattelaer, C.-A, Mohitosh, M, Smets, L, Maiti, M, Schepers, G, Mattelaer, H.-P, Rosemeyer, H, Herdewijn, P, Lescrinier, E. | | Deposit date: | 2021-01-04 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Stable Hairpin Structures Formed by Xylose-Based Nucleic Acids.
Chembiochem, 22, 2021
|
|
