1HL8
 
 | |
2V4V
 
 | | Crystal Structure of a Family 6 Carbohydrate-Binding Module from Clostridium cellulolyticum in complex with xylose | | Descriptor: | GH59 GALACTOSIDASE, SODIUM ION, beta-D-xylopyranose | | Authors: | Abbott, D.W, Ficko-Blean, E, Lammerts van Bueren, A, Coutinho, P.M, Henrissat, B, Gilbert, H.J, Boraston, A.B. | | Deposit date: | 2008-09-29 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of the Structural and Functional Diversity of Plant Cell Wall Specific Family 6 Carbohydrate Binding Modules.
Biochemistry, 48, 2009
|
|
4MAH
 
 | | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn | | Descriptor: | 1,2-ETHANEDIOL, AA11 Lytic Polysaccharide Monooxygenase, CHLORIDE ION, ... | | Authors: | Hemsworth, G.R, Henrissat, B, Walton, P.H, Davies, G.J. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and characterization of a new family of lytic polysaccharide monooxygenases.
Nat.Chem.Biol., 10, 2014
|
|
4MAI
 
 | | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) | | Descriptor: | 1,2-ETHANEDIOL, AA11 Lytic Polysaccharide Monooxygenase, CHLORIDE ION, ... | | Authors: | Hemsworth, G.R, Henrissat, B, Walton, P.H, Davies, G.J. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and characterization of a new family of lytic polysaccharide monooxygenases.
Nat.Chem.Biol., 10, 2014
|
|
4URI
 
 | | Crystal structure of chitinase-like agglutinin RobpsCRA from Robinia pseudoacacia | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHITINASE-RELATED AGGLUTININ, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Henrissat, B, van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2014-06-30 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Carbohydrate Binding Properties of a Plant Chitinase-Like Agglutinin with Conserved Catalytic Machinery.
J.Struct.Biol., 190, 2015
|
|
4UFC
 
 | | Crystal structure of the GH95 enzyme BACOVA_03438 | | Descriptor: | CACODYLATE ION, CALCIUM ION, GH95, ... | | Authors: | Rogowski, A, Briggs, J.A, Mortimer, J.C, Tryfona, T, Terrapon, N, Lowe, E.C, Basle, A, Morland, C, Day, A.M, Zheng, H, Rogers, T.E, Thompson, P, Hawkins, A.R, Yadav, M.P, Henrissat, B, Martens, E.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Glycan Complexity Dictates Microbial Resource Allocation in the Large Intestine.
Nat.Commun., 6, 2015
|
|
3W5M
 
 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Putative rhamnosidase | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
3W5N
 
 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase complexed with L-rhamnose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Putative rhamnosidase, ... | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
2BS9
 
 | | Native crystal structure of a GH39 beta-xylosidase XynB1 from Geobacillus stearothermophilus | | Descriptor: | BETA-XYLOSIDASE, CALCIUM ION | | Authors: | Czjzek, M, Bravman, T, Henrissat, B, Shoham, Y. | | Deposit date: | 2005-05-19 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzyme-Substrate Complex Structures of a Gh39 Beta-Xylosidase from Geobacillus Stearothermophilus.
J.Mol.Biol., 353, 2005
|
|
4OPB
 
 | | AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Lo Leggio, L, Frandsen, K.H, Davies, G.J, Dupree, P, Walton, P, Henrissat, B, Stringer, M, Tovborg, M, De Maria, L, Johansen, K.S. | | Deposit date: | 2014-02-05 | | Release date: | 2015-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase.
Nat Commun, 6, 2015
|
|
2BFG
 
 | | crystal structure of beta-xylosidase (fam GH39) in complex with dinitrophenyl-beta-xyloside and covalently bound xyloside | | Descriptor: | 2,5-DINITROPHENOL, BETA-XYLOSIDASE, SODIUM ION, ... | | Authors: | Czjzek, M, Bravman, T, Henrissat, B, Shoham, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzyme-Substrate Complex Structures of a Gh39 Beta-Xylosidase from Geobacillus Stearothermophilus.
J.Mol.Biol., 353, 2005
|
|
6I1T
 
 | | Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Turbe-Doan, A, Record, E, Lombard, V, Kumar, R, Henrissat, B, Levasseur, A, Garron, M.L. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The functional and structural characterization ofTrichoderma reeseidehydrogenase belonging to the PQQ dependent family of Carbohydrate-Active Enzymes Family AA12.
Appl.Environ.Microbiol., 2019
|
|
6I1Q
 
 | | Iodide structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Turbe-Doan, A, Record, E, Lombard, V, Kumar, R, Henrissat, B, Levasseur, A, Garron, M.L. | | Deposit date: | 2018-10-30 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The functional and structural characterization ofTrichoderma reeseidehydrogenase belonging to the PQQ dependent family of Carbohydrate-Active Enzymes Family AA12.
Appl.Environ.Microbiol., 2019
|
|
5C86
 
 | | Novel fungal alcohol oxidase with catalytic diversity among the AA5 family, apo form | | Descriptor: | Kelch domain-containing protein | | Authors: | Urresti, S, Yin, D.T, LaFond, M, Derikvand, F, Berrin, G.J, Henrissat, B, Walton, P.H, Brumer, H, Davies, G.J. | | Deposit date: | 2015-06-25 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family.
Nat Commun, 6, 2015
|
|
6FHN
 
 | | Structural dynamics and catalytic properties of a multi-modular xanthanase (Pt derivative) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
6FHJ
 
 | | Structural dynamics and catalytic properties of a multi-modular xanthanase, native. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
5G2U
 
 | | Structure of BT1596,a 2-O GAG sulfatase | | Descriptor: | 2-O GLYCOSAMINOGLYCAN SULFATASE, CITRIC ACID, ZINC ION | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G2V
 
 | | Structure of BT4656 in complex with its substrate D-Glucosamine-2-N, 6-O-disulfate. | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, CALCIUM ION, N-ACETYLGLUCOSAMINE-6-SULFATASE, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4B1L
 
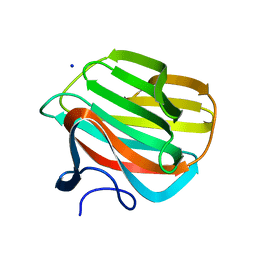 | | CARBOHYDRATE BINDING MODULE CBM66 FROM BACILLUS SUBTILIS | | Descriptor: | LEVANASE, SODIUM ION, beta-D-fructofuranose | | Authors: | Cuskin, F, Flint, J.E, Morland, C, Basle, A, Henrissat, B, Countinho, P.M, Strazzulli, A, Solzehinkin, A, Davies, G.J, Gilbert, H.J, Gloster, T.M. | | Deposit date: | 2012-07-11 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | How Nature Can Exploit Nonspecific Catalytic and Carbohydrate Binding Modules to Create Enzymatic Specificity
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1HL9
 
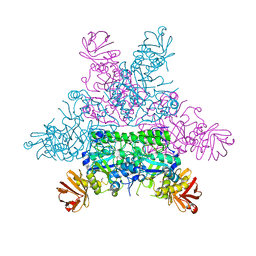 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA ALPHA-FUCOSIDASE IN COMPLEX WITH A MECHANISM BASED INHIBITOR | | Descriptor: | 2-deoxy-2-fluoro-beta-L-fucopyranose, PUTATIVE ALPHA-L-FUCOSIDASE | | Authors: | Sulzenbacher, G, Bignon, C, Bourne, Y, Henrissat, B. | | Deposit date: | 2003-03-14 | | Release date: | 2004-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Thermotoga Maritima {Alpha}-L-Fucosidase: Insights Into the Catalytic Mechanism and the Molecular Basis for Fucosidosis
J.Biol.Chem., 279, 2004
|
|
5C92
 
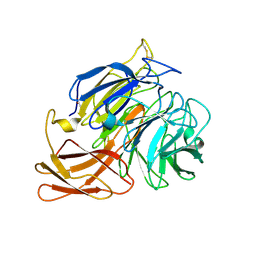 | | Novel fungal alcohol oxidase with catalytic diversity among the AA5 family, in complex with copper | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (I) ION, ... | | Authors: | Urresti, S, Yin, D.T, LaFond, M, Derikvand, F, Berrin, G.J, Henrissat, B, Walton, P.H, Brumer, H, Davies, G.J. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family.
Nat Commun, 6, 2015
|
|
4B1M
 
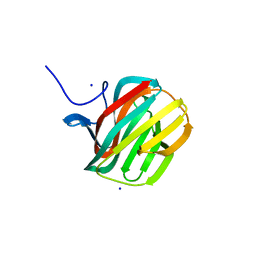 | | CARBOHYDRATE BINDING MODULE CBM66 FROM BACILLUS SUBTILIS | | Descriptor: | LEVANASE, SODIUM ION, SULFATE ION, ... | | Authors: | Cuskin, F, Flint, J.E, Morland, C, Basle, A, Henrissat, B, Countinho, P.M, Strazzulli, A, Solzehinkin, A, Davies, G.J, Gilbert, H.J, Gloster, T.M. | | Deposit date: | 2012-07-11 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | How Nature Can Exploit Nonspecific Catalytic and Carbohydrate Binding Modules to Create Enzymatic Specificity
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5G2T
 
 | | BT1596 in complex with its substrate 4,5 unsaturated uronic acid alpha 1,4 D-Glucosamine-2-N, 6-O-disulfate | | Descriptor: | 1,2-ETHANEDIOL, 2-O GLYCOSAMINOGLYCAN SULFATASE, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid, ... | | Authors: | Cartmell, A, Lowe, E.C, Basle, A, Crouch, L.I, Czjzek, M, Turnbull, J, Henrissat, B, Terrapon, N, Thomas, S, Murray, H, Firbank, S.J, Bolam, D.N. | | Deposit date: | 2016-04-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1ODU
 
 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA ALPHA-FUCOSIDASE IN COMPLEX WITH FUCOSE | | Descriptor: | PUTATIVE ALPHA-L-FUCOSIDASE, beta-L-fucopyranose | | Authors: | Sulzenbacher, G, Bignon, C, Bourne, Y, Henrissat, B. | | Deposit date: | 2003-03-14 | | Release date: | 2004-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Thermotoga Maritima {Alpha}-L-Fucosidase: Insights Into the Catalytic Mechanism and the Molecular Basis for Fucosidosis
J.Biol.Chem., 279, 2004
|
|
6H7T
 
 | | Native structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Turbe-Doan, A, Record, E, Lombard, V, Kumar, R, Henrissat, B, Levasseur, A, Garron, M.L. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The functional and structural characterization of Trichoderma reesei dehydrogenase belonging to the PQQ dependent family of Carbohydrate-Active Enzymes Family AA12.
Appl.Environ.Microbiol., 2019
|
|
