6A7B
 
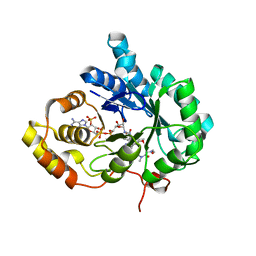 | | AKR1C3 complexed with new inhibitor with novel scaffold | | Descriptor: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C3, DIMETHYLFORMAMIDE, ... | | Authors: | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
6JIF
 
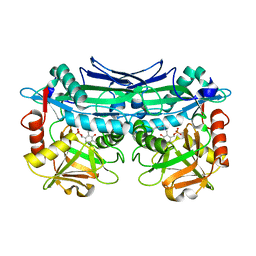 | | Crystal Structures of Branched-Chain Aminotransferase from Pseudomonas sp. UW4 | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zheng, X, Guo, L, Li, D.F, Wu, B. | | Deposit date: | 2019-02-21 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of a highly active branched-chain amino acid aminotransferase from Pseudomonas sp. for efficient biosynthesis of chiral amino acids.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
5ZXF
 
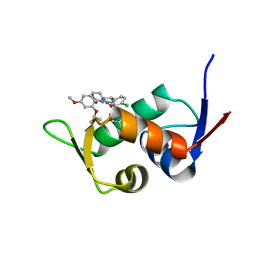 | | The 1.25A Crystal structure of His6-tagged Mdm2 in complex with nutlin-3a | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Cheng, X.Y, Su, Z.D, Pi, N, Cao, C.Z, Zhao, Z.T, Zhou, J.J, Chen, R, Kuang, Z.K, Huang, Y.Q. | | Deposit date: | 2018-05-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.25A Crystal structure of His6-tagged Mdm2 in complex with nutlin-3a
To Be Published
|
|
8J44
 
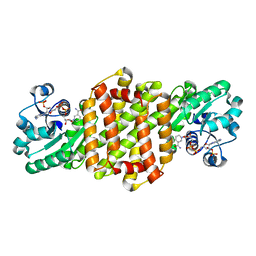 | | Reductive Aminase RA34-WT | | Descriptor: | 6-phosphogluconate dehydrogenase NAD-binding protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.Y, Xu, G.C, Ni, Y. | | Deposit date: | 2023-04-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dynamic kinetic reductive resolution of cyclic keto esters by newly identified stereo complementary reductive aminases.
To Be Published
|
|
8J43
 
 | | Reductive aminase RA29-WT | | Descriptor: | 1,2-ETHANEDIOL, Beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate dehydrogenase, ... | | Authors: | Zheng, X.Y, Xu, G.C, Ni, Y. | | Deposit date: | 2023-04-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Dynamic kinetic reductive resolution of cyclic keto esters by newly identified stereo complementary reductive aminases.
To Be Published
|
|
2DDY
 
 | | Solution Structure of Matrilysin (MMP-7) Complexed to Constraint Conformational Sulfonamide Inhibitor | | Descriptor: | (1R)-N,6-DIHYDROXY-7-METHOXY-2-[(4-METHOXYPHENYL)SULFONYL]-1,2,3,4-TETRAHYDROISOQUINOLINE-1-CARBOXAMIDE, CALCIUM ION, Matrilysin, ... | | Authors: | Zheng, X.H, Ou, L. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Matrilysin (MMP-7) Complexed to Constraint Conformational Sulfonamide Inhibitor
to be published
|
|
7C44
 
 | | Crystal structure of the p53-binding domain of human MdmX protein in complex with Nutlin3a | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, Protein Mdm4 | | Authors: | Cheng, X.Y, Zhang, B.L, Kuang, Z.K, Yang, J, Li, Z.C, Yu, J.P, Zhao, Z.T, Cao, C.Z, Su, Z.D. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the p53-binding domain of human MdmX protein in complex with Nutlin3a
To Be Published
|
|
7EL4
 
 | | The crystal structure of p53p peptide fragment in complex with the N-terminal domain of MdmX | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Cellular tumor antigen p53,Protein Mdm4, MAGNESIUM ION | | Authors: | Cheng, X.Y, Zhang, B.L, Li, Z.C, Kuang, Z.K, Su, Z.D. | | Deposit date: | 2021-04-08 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The crystal structure of the N-terminal domain of MdmX in complex with p53p peptide fragment
To Be Published
|
|
5CX3
 
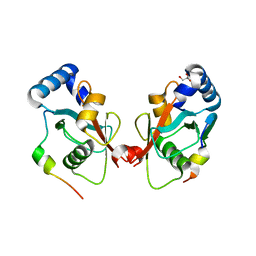 | | Crystal structure of FYCO1 LIR in complex with LC3A | | Descriptor: | FYVE and coiled-coil domain-containing protein 1, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Cheng, X, Pan, L. | | Deposit date: | 2015-07-28 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of FYCO1 and MAP1LC3A interaction reveals a novel binding mode for Atg8-family proteins.
Autophagy, 12, 2016
|
|
6KAC
 
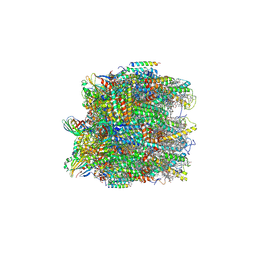 | | Cryo-EM structure of the C2S2-type PSII-LHCII supercomplex from Chlamydomonas reihardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Sheng, X, Li, A.J, Song, D.F, Liu, Z.F. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-23 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insight into light harvesting for photosystem II in green algae.
Nat.Plants, 5, 2019
|
|
6KAD
 
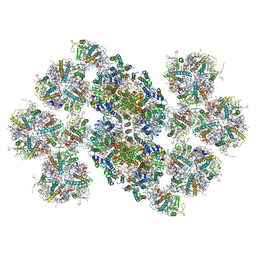 | | Cryo-EM structure of the C2S2M2L2-type PSII-LHCII supercomplex from Chlamydomonas reihardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Sheng, X, Watanabe, A, Li, A.J, Kim, E, Song, C, Murata, K, Song, D.F, Minagawa, J, Liu, Z.F. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insight into light harvesting for photosystem II in green algae.
Nat.Plants, 5, 2019
|
|
7CGL
 
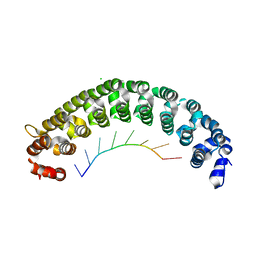 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5C (5'-R(P*UP*GP*UP*AP*CP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGI
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5U (5'-R(P*UP*GP*UP*AP*UP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGF
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5G (5'-R(P*UP*GP*UP*AP*GP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGG
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5A (5'-R(P*UP*GP*UP*AP*AP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGH
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5C (5'-R(P*UP*GP*UP*AP*CP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGK
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5A (5'-R(P*UP*GP*UP*AP*AP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGM
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5U (5'-R(P*UP*GP*UP*AP*UP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7CGJ
 
 | | Crystal Structure of PUF-8 in Complex with PBE-RNA | | Descriptor: | CHLORIDE ION, PBE-5G (5'-R(P*UP*GP*UP*AP*GP*AP*UP*A)-3'), PUM-HD domain-containing protein | | Authors: | Zheng, X, Yunyu, S, Shouhong, G. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.546 Å) | | Cite: | Structural recognition of the mRNA 3' UTR by PUF-8 restricts the lifespan of C. elegans.
Nucleic Acids Res., 49, 2021
|
|
7RWR
 
 | | An RNA aptamer that decreases flavin redox potential | | Descriptor: | FLAVIN MONONUCLEOTIDE, RNA (38-MER) | | Authors: | Gremminger, T, Li, J, Chen, S, Heng, X. | | Deposit date: | 2021-08-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An RNA aptamer that shifts the reduction potential of metabolic cofactors.
Nat.Chem.Biol., 18, 2022
|
|
4LT5
 
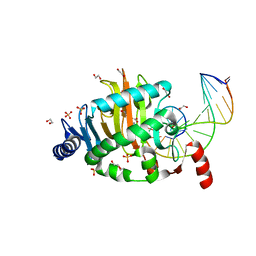 | | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA, MANGANESE (II) ION, ... | | Authors: | Hashimoto, H, Pais, J.E, Zhang, X, Saleh, L, Fu, Z.Q, Dai, N, Correa, I.R, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2013-07-23 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.
Nature, 506, 2014
|
|
1YFZ
 
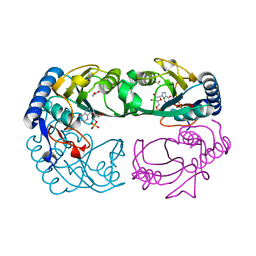 | | Novel IMP Binding in Feedback Inhibition of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis | | Descriptor: | ACETATE ION, Hypoxanthine-guanine phosphoribosyltransferase, INOSINIC ACID, ... | | Authors: | Chen, Q, Liang, Y, Su, X, Gu, X, Zheng, X, Luo, M. | | Deposit date: | 2005-01-04 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alternative IMP Binding in Feedback Inhibition of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis.
J.Mol.Biol., 348, 2005
|
|
1YFJ
 
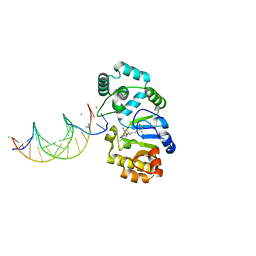 | | T4Dam in Complex with AdoHcy and 15-mer Oligonucleotide Showing Semi-specific and Specific Contact | | Descriptor: | 5'-D(*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*G)-3', CALCIUM ION, CHLORIDE ION, ... | | Authors: | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | Deposit date: | 2005-01-02 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
6DQB
 
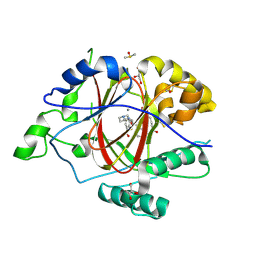 | | LINKED KDM5A JMJ DOMAIN FORMING COVALENT BOND TO INHIBITOR N71 i.e. 2-((3-(4-(dimethylamino)but-2-enamido)phenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 2-{(R)-(3-{[(2E)-4-(dimethylamino)but-2-enoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(R)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(S)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ8
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR N49 i.e. 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-[(R)-(2-chlorophenyl){2-[(2S)-1-methylpyrrolidin-2-yl]ethoxy}methyl]thieno[3,2-b]pyridine-7-carboxylic acid, 2-[(S)-(2-chlorophenyl){2-[(2S)-1-methylpyrrolidin-2-yl]ethoxy}methyl]thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
