2BRD
 
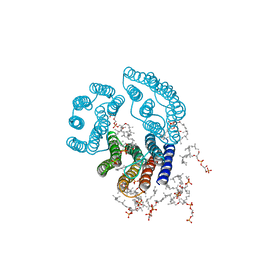 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN PURPLE MEMBRANE | | Descriptor: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | Authors: | Henderson, R, Grigorieff, N. | | Deposit date: | 1995-12-27 | | Release date: | 1996-06-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Electron-crystallographic refinement of the structure of bacteriorhodopsin.
J.Mol.Biol., 259, 1996
|
|
1BRD
 
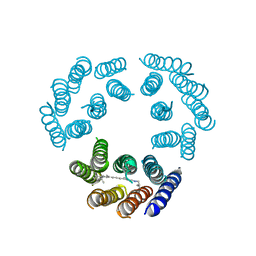 | | Model for the structure of Bacteriorhodopsin based on high-resolution Electron Cryo-microscopy | | Descriptor: | BACTERIORHODOPSIN PRECURSOR, RETINAL | | Authors: | Henderson, R, Baldwin, J.M, Ceska, T.A, Zemlin, F, Beckmann, E, Downing, K.H. | | Deposit date: | 1990-05-23 | | Release date: | 1991-04-15 | | Last modified: | 2024-04-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy.
J.Mol.Biol., 213, 1990
|
|
6X2A
 
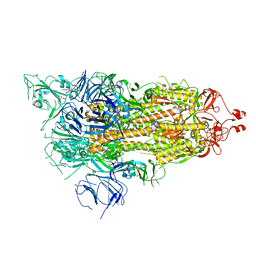 | |
6X2B
 
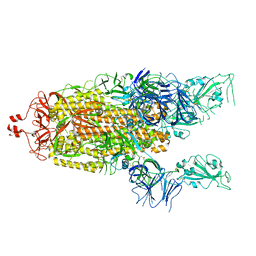 | |
6X29
 
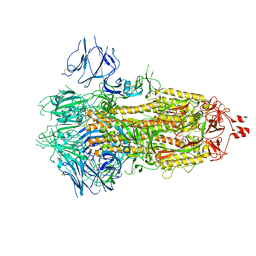 | |
6X2C
 
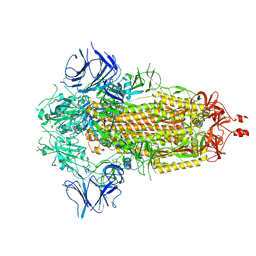 | |
7M0J
 
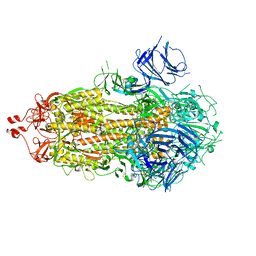 | |
6V8Z
 
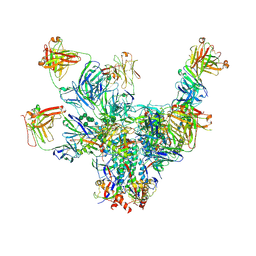 | |
6V8X
 
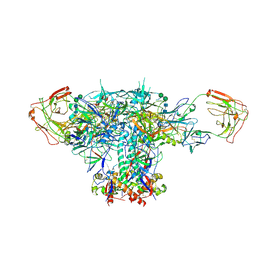 | | VRC01 Bound BG505 F14 HIV-1 SOSIP Envelope Trimer Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2019-12-12 | | Release date: | 2020-02-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Disruption of the HIV-1 Envelope allosteric network blocks CD4-induced rearrangements.
Nat Commun, 11, 2020
|
|
6UDA
 
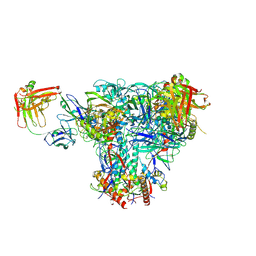 | | Cryo-EM structure of CH235UCA bound to Man5-enriched CH505.N279K.G458Y.SOSIP.664 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH235 UCA heavy chain Fab, CH235 UCA light chain Fab, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2019-09-19 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancester of CH235 lineage CD4bs broadly neutralizing antibodies.
Plos Pathog., 15, 2019
|
|
7TLA
 
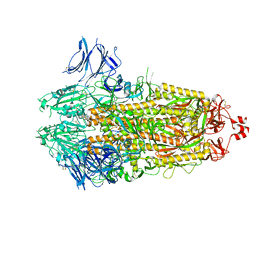 | |
7TLD
 
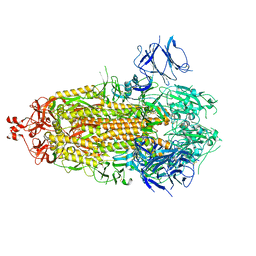 | |
7TLB
 
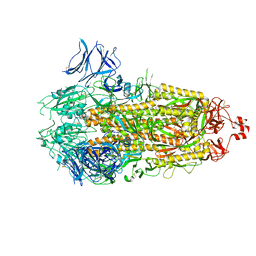 | |
7TLC
 
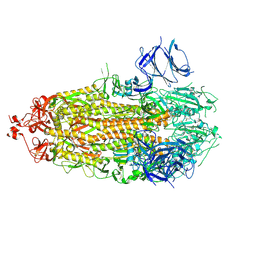 | |
8SXJ
 
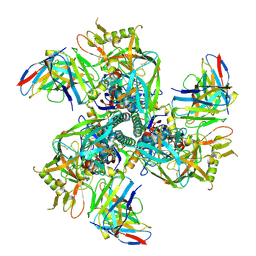 | | CH505 Disulfide Stapled SOSIP Bound to CH235.12 Fab | | Descriptor: | CH235.12 Heavy Chain, CH235.12 Light Chain, Envelope glycoprotein gp160, ... | | Authors: | Henderson, R. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv, 10, 2024
|
|
8SXI
 
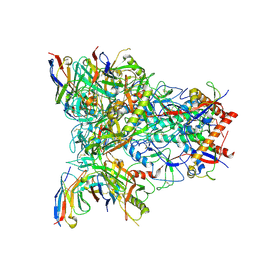 | | CH505 Disulfide Stapled SOSIP Bound to b12 Fab | | Descriptor: | Envelope glycoprotein gp160, HIV-1 gp41, b12 Heavy Chain, ... | | Authors: | Henderson, R. | | Deposit date: | 2023-05-22 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Microsecond dynamics control the HIV-1 Envelope conformation.
Sci Adv, 10, 2024
|
|
8SAW
 
 | | CryoEM structure of DH270.UCA.G57R-CH848.10.17DT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.3.D0949.10.17chim.6R.SOSIP.664 gp120A, CH848.3.D0949.10.17chim.6R.SOSIP.664 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAN
 
 | | CryoEM structure of VRC01-CH848.0836.10 | | Descriptor: | CH848.0836.10 gp120, CH848.0836.10 gp41, VCR01 variable heavy chain, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAU
 
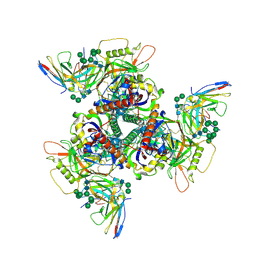 | | CryoEM structure of DH270.4-CH848.10.17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17 gp120, CH848.10.17 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAY
 
 | | CryoEM structure of DH270.3-CH848.10.17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17 gp120, CH848.10.17 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAR
 
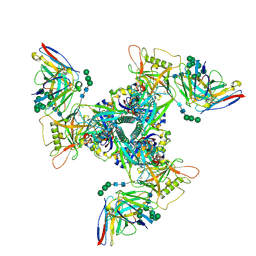 | | CryoEM structure of DH270.6-CH848.10.17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17 gp120, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SB3
 
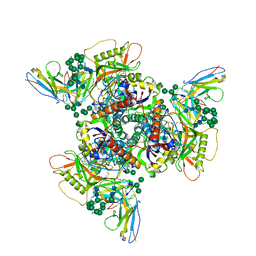 | | CryoEM structure of DH270.2-CH848.10.17 | | Descriptor: | CH848.10.17 gp120, CH848.10.17.SOSIP gp41, DH270.2 Variable Heavy chain, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAV
 
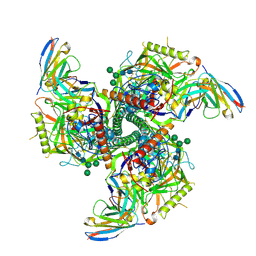 | | CryoEM structure of VRC01-CH848.0526.25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.0526.25 gp120, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAX
 
 | | CryoEM structure of DH270.UCA-CH848.10.17DT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17.SOSIP gp120, CH848.10.17.SOSIP gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAZ
 
 | | CryoEM structure of DH270.I5.6-CH848.10.17 | | Descriptor: | CH848.10.17.SOSIP gp41, DH270.I5.6 variable heavy chain, DH270.I5.6 variable light chain, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
