4XQF
 
 | | Tailspike protein mutant E372Q (delta D470/N471) of E. coli bacteriophage HK620 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-19 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4YEJ
 
 | | Tailspike protein double mutant D339A/E372Q of E. coli bacteriophage HK620 in complex with pentasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4YEL
 
 | | Tailspike protein double mutant D339A/E372A of E. coli bacteriophage HK620 in complex with hexasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
2I5L
 
 | |
2I5M
 
 | |
1UN2
 
 | | Crystal structure of circularly permuted CPDSBA_Q100T99: Preserved Global Fold and Local Structural Adjustments | | Descriptor: | THIOL-DISULFIDE INTERCHANGE PROTEIN | | Authors: | Manjasetty, B.A, Hennecke, J, Glockshuber, R, Heinemann, U. | | Deposit date: | 2003-09-03 | | Release date: | 2003-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Circularly Permuted Dsba(Q100T99): Preserved Global Fold and Local Structural Adjustments
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4XNF
 
 | | Tailspike protein double mutant D339A/E372Q of E. coli bacteriophage HK620 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
1U06
 
 | | crystal structure of chicken alpha-spectrin SH3 domain | | Descriptor: | AZIDE ION, Spectrin alpha chain, brain | | Authors: | Chevelkov, V, Faelber, K, Diehl, A, Heinemann, U, Oschkinat, H, Reif, B. | | Deposit date: | 2004-07-13 | | Release date: | 2005-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Detection of dynamic water molecules in a microcrystalline sample of the SH3 domain of alpha-spectrin by MAS solid-state NMR.
J.Biomol.Nmr, 31, 2005
|
|
1U0A
 
 | | Crystal structure of the engineered beta-1,3-1,4-endoglucanase H(A16-M) in complex with beta-glucan tetrasaccharide | | Descriptor: | Beta-glucanase, CALCIUM ION, ZINC ION, ... | | Authors: | Gaiser, O.J, Piotukh, K, Ponnuswamy, M.N, Planas, A, Borriss, R, Heinemann, U. | | Deposit date: | 2004-07-13 | | Release date: | 2005-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Basis for the Substrate Specificity of a Bacillus 1,3-1,4-beta-Glucanase
J.Mol.Biol., 357, 2006
|
|
1ONI
 
 | | Crystal structure of a human p14.5, a translational inhibitor reveals different mode of ligand binding near the invariant residues of the Yjgf/UK114 protein family | | Descriptor: | 14.5 kDa translational inhibitor protein, BENZOIC ACID | | Authors: | Manjasetty, B.A, Delbrueck, H, Mueller, U, Erdmann, M.F, Heinemann, U. | | Deposit date: | 2003-02-28 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Homo sapiens protein hp14.5.
Proteins, 54, 2004
|
|
3TT9
 
 | | Crystal structure of the stable degradation fragment of human plakophilin 2 isoform a (PKP2a) C752R variant | | Descriptor: | GLYCEROL, Plakophilin-2 | | Authors: | Schuetz, A, Roske, Y, Gerull, B, Heinemann, U. | | Deposit date: | 2011-09-14 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular insights into arrhythmogenic right ventricular cardiomyopathy caused by plakophilin-2 missense mutations.
Circ Cardiovasc Genet, 5, 2012
|
|
3RIQ
 
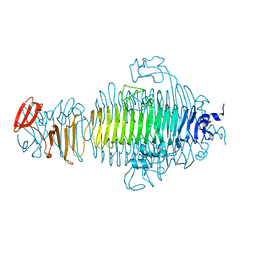 | | Siphovirus 9NA tailspike receptor binding domain | | Descriptor: | GLYCEROL, Tailspike protein | | Authors: | Andres, D, Roske, Y, Doering, C, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2011-04-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tail morphology controls DNA release in two Salmonella phages with one lipopolysaccharide receptor recognition system.
Mol.Microbiol., 83, 2012
|
|
1RGL
 
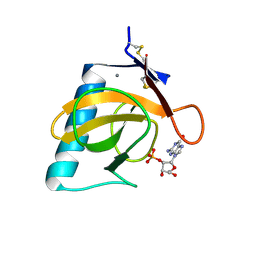 | | RNASE T1 MUTANT GLU46GLN BINDS THE INHIBITORS 2'GMP AND 2'AMP AT THE 3' SUBSITE | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Granzin, J, Puras-Lutzke, R, Landt, O, Grunert, H.-P, Heinemann, U, Saenger, W, Hahn, U. | | Deposit date: | 1992-02-19 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RNase T1 mutant Glu46Gln binds the inhibitors 2'GMP and 2'AMP at the 3' subsite.
J.Mol.Biol., 225, 1992
|
|
1RGK
 
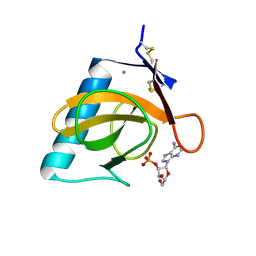 | | RNASE T1 MUTANT GLU46GLN BINDS THE INHIBITORS 2'GMP AND 2'AMP AT THE 3' SUBSITE | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, CALCIUM ION, RIBONUCLEASE T1 | | Authors: | Granzin, J, Puras-Lutzke, R, Landt, O, Grunert, H.-P, Heinemann, U, Saenger, W, Hahn, U. | | Deposit date: | 1992-02-19 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | RNase T1 mutant Glu46Gln binds the inhibitors 2'GMP and 2'AMP at the 3' subsite.
J.Mol.Biol., 225, 1992
|
|
3ULJ
 
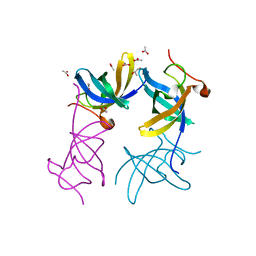 | | Crystal structure of apo Lin28B cold shock domain | | Descriptor: | ACETATE ION, GLYCEROL, Lin28b, ... | | Authors: | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | Deposit date: | 2011-11-10 | | Release date: | 2012-08-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The Lin28 cold-shock domain remodels pre-let-7 microRNA.
Nucleic Acids Res., 40, 2012
|
|
2R80
 
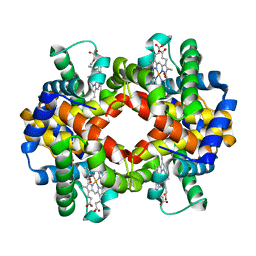 | | Pigeon Hemoglobin (OXY form) | | Descriptor: | Hemoglobin subunit alpha-A, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Ponnuswamy, M.N, Packianathan, C, Sundaresan, S, Neelagandan, K, Palani, K, Muller, J.J, Heinemann, U. | | Deposit date: | 2007-09-10 | | Release date: | 2008-09-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | X-ray crystal structure analysis of Hemolgobin from Pigeon (Columba Livia) at 1.44 angstrom
To be Published
|
|
1AQ0
 
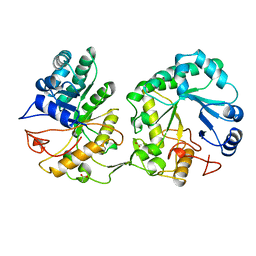 | | BARLEY 1,3-1,4-BETA-GLUCANASE IN MONOCLINIC SPACE GROUP | | Descriptor: | 1,3-1,4-BETA-GLUCANASE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION | | Authors: | Mueller, J.J, Thomsen, K.K, Heinemann, U. | | Deposit date: | 1997-08-05 | | Release date: | 1998-02-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of barley 1,3-1,4-beta-glucanase at 2.0-A resolution and comparison with Bacillus 1,3-1,4-beta-glucanase.
J.Biol.Chem., 273, 1998
|
|
1B67
 
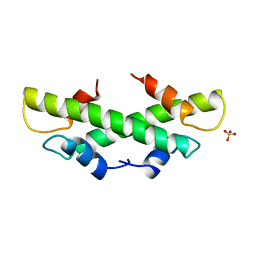 | | CRYSTAL STRUCTURE OF THE HISTONE HMFA FROM METHANOTHERMUS FERVIDUS | | Descriptor: | PROTEIN (HISTONE HMFA), SULFATE ION | | Authors: | Decanniere, K, Sandman, K, Reeve, J.N, Heinemann, U. | | Deposit date: | 1999-01-19 | | Release date: | 2000-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus.
J.Mol.Biol., 303, 2000
|
|
1B6W
 
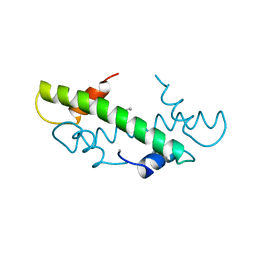 | |
1A7W
 
 | | CRYSTAL STRUCTURE OF THE HISTONE HMFB FROM METHANOTHERMUS FERVIDUS | | Descriptor: | CHLORIDE ION, HISTONE HMFB, ZINC ION | | Authors: | Decanniere, K, Sandman, K, Reeve, J.N, Heinemann, U. | | Deposit date: | 1998-03-18 | | Release date: | 1999-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus.
J.Mol.Biol., 303, 2000
|
|
1GBG
 
 | | BACILLUS LICHENIFORMIS BETA-GLUCANASE | | Descriptor: | (1,3-1,4)-BETA-D-GLUCAN 4 GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1995-08-25 | | Release date: | 1995-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus licheniformis 1,3-1,4-beta-D-glucan 4-glucanohydrolase at 1.8 A resolution.
FEBS Lett., 374, 1995
|
|
3O5N
 
 | | Tetrahydroquinoline carboxylates are potent inhibitors of the Shank PDZ domain, a putative target in autism disorders | | Descriptor: | (3aS,4R,9bR)-9-nitro-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-4,6-dicarboxylic acid, SH3 and multiple ankyrin repeat domains protein 3 | | Authors: | Saupe, J, Roske, Y, Schillinger, C, Kamdem, N, Radetzki, S, Diehl, A, Oschkinat, H, Krause, G, Heinemann, U, Rademann, J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery, structure-activity relationship studies, and crystal structure of nonpeptide inhibitors bound to the shank3 PDZ domain.
Chemmedchem, 6, 2011
|
|
3PF4
 
 | | Crystal structure of Bs-CspB in complex with r(GUCUUUA) | | Descriptor: | Cold shock protein cspB, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Sachs, R, Max, K.E.A, Heinemann, U. | | Deposit date: | 2010-10-27 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution.
Rna, 18, 2012
|
|
3PF5
 
 | |
3PDO
 
 | | Crystal Structure of HLA-DR1 with CLIP102-120 | | Descriptor: | FORMIC ACID, GLYCEROL, HLA class II histocompatibility antigen gamma chain, ... | | Authors: | Gunther, S, Schlundt, A, Sticht, J, Roske, Y, Heinemann, U, Wiesmuller, K.-H, Jung, G, Falk, K, Rotzschke, O, Freund, C. | | Deposit date: | 2010-10-23 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bidirectional binding of invariant chain peptides to an MHC class II molecule.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
