5IFW
 
 | |
5IFS
 
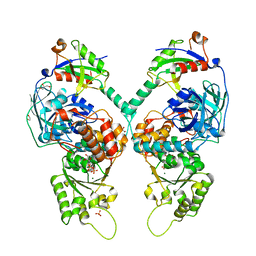 | | Quantitative interaction mapping reveals an extended ubiquitin regulatory domain in ASPL that disrupts functional p97 hexamers and induces cell death | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Roske, Y, Arumughan, A, Heinemann, U, Wanker, E. | | Deposit date: | 2016-02-26 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Quantitative interaction mapping reveals an extended UBX domain in ASPL that disrupts functional p97 hexamers.
Nat Commun, 7, 2016
|
|
1GBG
 
 | | BACILLUS LICHENIFORMIS BETA-GLUCANASE | | Descriptor: | (1,3-1,4)-BETA-D-GLUCAN 4 GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1995-08-25 | | Release date: | 1995-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus licheniformis 1,3-1,4-beta-D-glucan 4-glucanohydrolase at 1.8 A resolution.
FEBS Lett., 374, 1995
|
|
3TH0
 
 | | P22 Tailspike complexed with S.Paratyphi O antigen octasaccharide | | Descriptor: | Bifunctional tail protein, GLYCEROL, alpha-D-galactopyranose-(1-2)-[alpha-D-Paratopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-alpha-D-galactopyranose-(1-2)-[alpha-D-Paratopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose | | Authors: | Andres, D, Gohlke, U, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2011-08-18 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An essential serotype recognition pocket on phage P22 tailspike protein forces Salmonella enterica serovar Paratyphi A O-antigen fragments to bind as nonsolution conformers.
Glycobiology, 23, 2013
|
|
1HTA
 
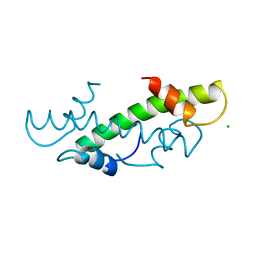 | | CRYSTAL STRUCTURE OF THE HISTONE HMFA FROM METHANOTHERMUS FERVIDUS | | Descriptor: | CHLORIDE ION, HISTONE HMFA | | Authors: | Decanniere, K, Sandman, K, Reeve, J.N, Heinemann, U. | | Deposit date: | 1998-03-18 | | Release date: | 1999-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of recombinant histones HMfA and HMfB from the hyperthermophilic archaeon Methanothermus fervidus.
J.Mol.Biol., 303, 2000
|
|
1ONI
 
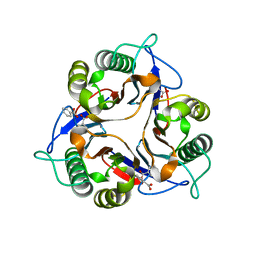 | | Crystal structure of a human p14.5, a translational inhibitor reveals different mode of ligand binding near the invariant residues of the Yjgf/UK114 protein family | | Descriptor: | 14.5 kDa translational inhibitor protein, BENZOIC ACID | | Authors: | Manjasetty, B.A, Delbrueck, H, Mueller, U, Erdmann, M.F, Heinemann, U. | | Deposit date: | 2003-02-28 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Homo sapiens protein hp14.5.
Proteins, 54, 2004
|
|
6Q3V
 
 | |
2ONQ
 
 | |
7ZIH
 
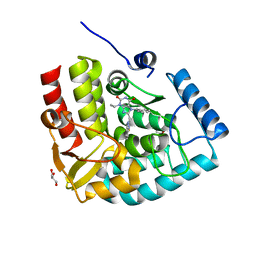 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor AG-01-128 | | Descriptor: | 8-(1~{H}-benzimidazol-2-ylmethyl)-3-ethyl-7-(phenylmethyl)purine-2,6-dione, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Schuetz, A, Gogolin, A, Pfeifer, J, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46890831 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIK
 
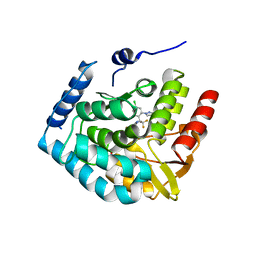 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor LP778902 | | Descriptor: | (2~{R})-2-azanyl-3-[4-[2-azanyl-6-[(1~{R})-1-[4-chloranyl-2-(3-methylpyrazol-1-yl)phenyl]-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]phenyl]propanoic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.58925915 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZII
 
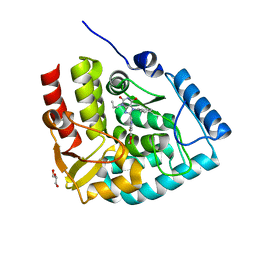 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-193 | | Descriptor: | 8-(5~{H}-[1,3]dioxolo[4,5-f]benzimidazol-6-ylmethyl)-7-(phenylmethyl)-3-propyl-purine-2,6-dione, FE (III) ION, GLYCEROL, ... | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6280005 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIJ
 
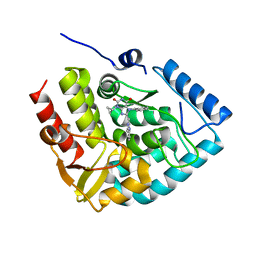 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-080 | | Descriptor: | 8-(1~{H}-benzimidazol-2-ylmethyl)-3-cyclopropyl-7-(phenylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94678366 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIG
 
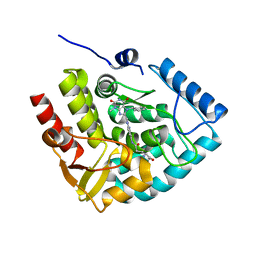 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-060 | | Descriptor: | (2~{R})-2-azanyl-5-[[2-[[3-methyl-2,6-bis(oxidanylidene)-7-(phenylmethyl)purin-8-yl]methyl]-1~{H}-benzimidazol-5-yl]amino]-5-oxidanylidene-pentanoic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.808885 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIF
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-480 | | Descriptor: | (2R)-2-azanyl-5-[[2-[3-methyl-2,6-bis(oxidanylidene)-7-(phenylmethyl)purin-8-yl]sulfanyl-3H-benzimidazol-5-yl]amino]-5-oxidanylidene-pentanoic acid, FE (III) ION, GLYCEROL, ... | | Authors: | Schuetz, A, Ziebart, N, Weise, M, Mallow, K, Pfeifer, J, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86859715 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
2OKN
 
 | | Crystal Strcture of Human Prolidase | | Descriptor: | HYDROGENPHOSPHATE ION, MANGANESE (II) ION, Xaa-Pro dipeptidase | | Authors: | Mueller, U, Niesen, F.H, Roske, Y, Goetz, F, Behlke, J, Buessow, K, Heinemann, U, Protein Structure Factory (PSF) | | Deposit date: | 2007-01-17 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Human Prolidase: The Molecular Basis of PD Disease.
To be Published
|
|
2ON8
 
 | |
2YIP
 
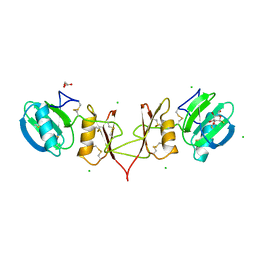 | |
1NEG
 
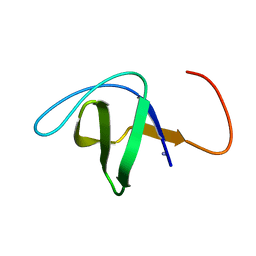 | | Crystal Structure Analysis of N-and C-terminal labeled SH3-domain of alpha-Chicken Spectrin | | Descriptor: | AZIDE ION, Spectrin alpha chain, brain | | Authors: | Mueller, U, Buessow, K, Diehl, A, Niesen, F.H, Nyarsik, L, Heinemann, U. | | Deposit date: | 2002-12-11 | | Release date: | 2003-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid purification and crystal structure analysis of a small protein carrying two terminal affinity tags
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
2YIL
 
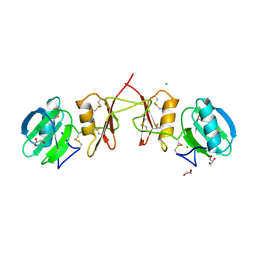 | | Crystal Structure of Parasite Sarcocystis muris Lectin SML-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, MICRONEME ANTIGEN L2, ... | | Authors: | Mueller, J.J, Weiss, M.S, Heinemann, U. | | Deposit date: | 2011-05-16 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pan-Modular Structure of Microneme Protein Sml-2 from Parasite Sarcocystis Muris at 1.95 A Resolution and its Complex with 1-Thio-Beta-D-Galactose.
Acta Crystallogr.,Sect.D, D67, 2011
|
|
1OQA
 
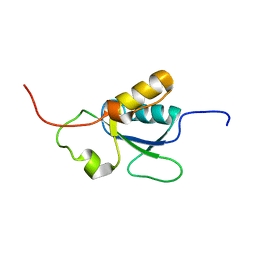 | | Solution structure of the BRCT-c domain from human BRCA1 | | Descriptor: | Breast cancer type 1 susceptibility protein | | Authors: | Gaiser, O.J, Ball, L.J, Schmieder, P, Leitner, D, Strauss, H, Wahl, M, Kuhne, R, Oschkinat, H, Heinemann, U. | | Deposit date: | 2003-03-07 | | Release date: | 2004-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics, and association behavior of the C-terminal BRCT domain from the breast cancer-associated protein BRCA1.
Biochemistry, 43, 2004
|
|
7NDJ
 
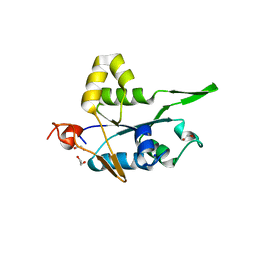 | |
1XCR
 
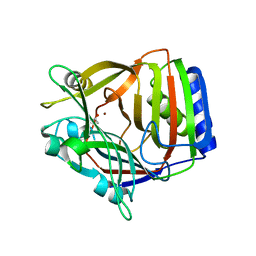 | | Crystal Structure of Longer Splice Variant of PTD012 from Homo sapiens reveals a novel Zinc-containing fold | | Descriptor: | ACETIC ACID, ZINC ION, hypothetical protein PTD012 | | Authors: | Manjasetty, B.A, Fieber-Erdmann, M, Roske, Y, Goetz, F, Buessow, K, Heinemann, U. | | Deposit date: | 2004-09-03 | | Release date: | 2005-09-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Homo sapiens PTD012 reveals a zinc-containing hydrolase fold
Protein Sci., 15, 2006
|
|
1OJH
 
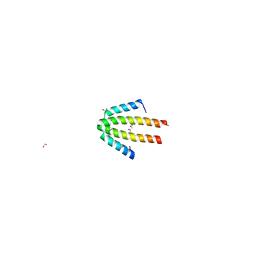 | | Crystal structure of NblA from PCC 7120 | | Descriptor: | 1,2-ETHANEDIOL, NBLA | | Authors: | Bienert, R, Baier, K, Lockau, W, Heinemann, U. | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Nbla from Anabaena Sp. Pcc 7120, a Small Protein Playing a Key Role in Phycobilisome Degradation.
J.Biol.Chem., 281, 2006
|
|
4XNF
 
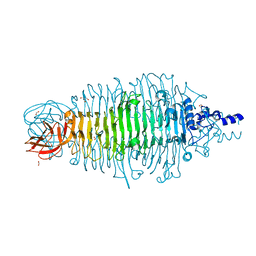 | | Tailspike protein double mutant D339A/E372Q of E. coli bacteriophage HK620 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XOP
 
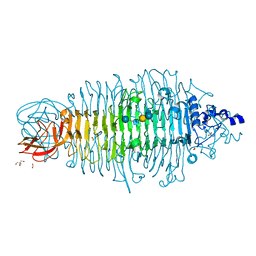 | | Tailspike protein double mutant D339N/E372Q of E. coli bacteriophage HK620 in complex with hexasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
