8CJO
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-06-004 | | Descriptor: | 1-[(3~{S})-3-(4-chloranyl-2-fluoranyl-phenyl)-1,4,8-triazatricyclo[7.4.0.0^{2,7}]trideca-2(7),8-dien-4-yl]-2-(2-ethyl-6-methyl-pyridin-3-yl)oxy-ethanone, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86633706 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
6XYD
 
 | | Crystal structure of Q4D6Q6, a conserved kinetoplastid-specific protein from Trypanosoma cruzi | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Roske, Y, Heinemann, U, Andrea, E.D. | | Deposit date: | 2020-01-30 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of Q4D6Q6, a conserved kinetoplastid-specific protein from Trypanosoma cruzi.
J.Struct.Biol., 211, 2020
|
|
8CJM
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-07-047 | | Descriptor: | 7-(cyclobutylmethyl)-3-ethyl-8-(5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-2-ylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8CJI
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-07-052 | | Descriptor: | FE (III) ION, Tryptophan 5-hydroxylase 1, methyl (2~{S})-2-azanyl-3-[[3-[[3-ethyl-2,6-bis(oxidanylidene)-8-(5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-2-ylmethyl)purin-7-yl]methyl]phenyl]carbonylamino]propanoate | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8CJJ
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-06-057 | | Descriptor: | 3-ethyl-7-(phenylmethyl)-8-(5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-2-ylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66415656 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8CJN
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-06-070 | | Descriptor: | 3-ethyl-7-[(4-phenylphenyl)methyl]-8-(5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-2-ylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68080938 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
8CJK
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-06-098 | | Descriptor: | 3-ethyl-8-[(2-methylimidazo[2,1-b][1,3]thiazol-6-yl)methyl]-7-[[4-(1-methylpyrazol-3-yl)phenyl]methyl]purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45914972 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
1E6E
 
 | | ADRENODOXIN REDUCTASE/ADRENODOXIN COMPLEX OF MITOCHONDRIAL P450 SYSTEMS | | Descriptor: | ADRENODOXIN, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mueller, J.J, Lapko, A, Bourenkov, G, Ruckpaul, K, Heinemann, U. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adrenodoxin Reductase-Adrenodoxin Complex Structure Suggests Electron Transfer Path in Steroid Biosynthesis.
J.Biol.Chem., 276, 2001
|
|
6HQC
 
 | |
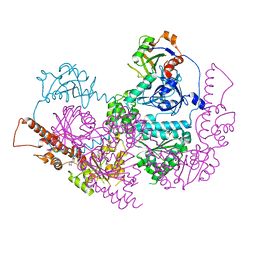
5IFW
 
 | |
1MJC
 
 | |
5IFS
 
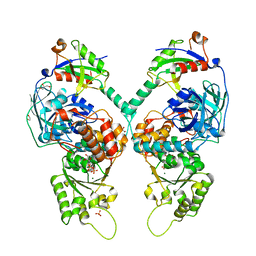 | | Quantitative interaction mapping reveals an extended ubiquitin regulatory domain in ASPL that disrupts functional p97 hexamers and induces cell death | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Roske, Y, Arumughan, A, Heinemann, U, Wanker, E. | | Deposit date: | 2016-02-26 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Quantitative interaction mapping reveals an extended UBX domain in ASPL that disrupts functional p97 hexamers.
Nat Commun, 7, 2016
|
|
1GLH
 
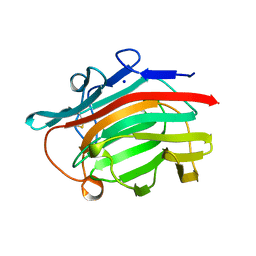 | | CATION BINDING TO A BACILLUS (1,3-1,4)-BETA-GLUCANASE. GEOMETRY, AFFINITY AND EFFECT ON PROTEIN STABILITY | | Descriptor: | 1,3-1,4-BETA-GLUCANASE, SODIUM ION | | Authors: | Keitel, T, Heinemann, U. | | Deposit date: | 1994-11-25 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cation binding to a Bacillus (1,3-1,4)-beta-glucanase. Geometry, affinity and effect on protein stability
Eur.J.Biochem., 222, 1994
|
|
7ZIK
 
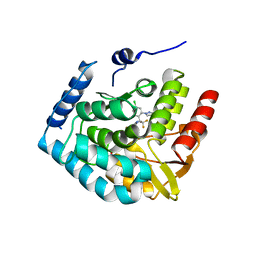 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor LP533401 | | Descriptor: | (2~{R})-2-azanyl-3-[4-[2-azanyl-6-[(1~{R})-1-[4-chloranyl-2-(3-methylpyrazol-1-yl)phenyl]-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]phenyl]propanoic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58925915 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIH
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor AG-01-128 | | Descriptor: | 8-(1~{H}-benzimidazol-2-ylmethyl)-3-ethyl-7-(phenylmethyl)purine-2,6-dione, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Schuetz, A, Gogolin, A, Pfeifer, J, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46890831 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZII
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-193 | | Descriptor: | 8-(5~{H}-[1,3]dioxolo[4,5-f]benzimidazol-6-ylmethyl)-7-(phenylmethyl)-3-propyl-purine-2,6-dione, FE (III) ION, GLYCEROL, ... | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6280005 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIJ
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-080 | | Descriptor: | 8-(1~{H}-benzimidazol-2-ylmethyl)-3-cyclopropyl-7-(phenylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94678366 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIG
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-060 | | Descriptor: | (2~{R})-2-azanyl-5-[[2-[[3-methyl-2,6-bis(oxidanylidene)-7-(phenylmethyl)purin-8-yl]methyl]-1~{H}-benzimidazol-5-yl]amino]-5-oxidanylidene-pentanoic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.808885 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIF
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-480 | | Descriptor: | (2R)-2-azanyl-5-[[2-[3-methyl-2,6-bis(oxidanylidene)-7-(phenylmethyl)purin-8-yl]sulfanyl-3H-benzimidazol-5-yl]amino]-5-oxidanylidene-pentanoic acid, FE (III) ION, GLYCEROL, ... | | Authors: | Schuetz, A, Ziebart, N, Weise, M, Mallow, K, Pfeifer, J, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86859715 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
3TH0
 
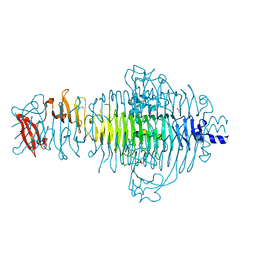 | | P22 Tailspike complexed with S.Paratyphi O antigen octasaccharide | | Descriptor: | Bifunctional tail protein, GLYCEROL, alpha-D-galactopyranose-(1-2)-[alpha-D-Paratopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-alpha-D-galactopyranose-(1-2)-[alpha-D-Paratopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose | | Authors: | Andres, D, Gohlke, U, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2011-08-18 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An essential serotype recognition pocket on phage P22 tailspike protein forces Salmonella enterica serovar Paratyphi A O-antigen fragments to bind as nonsolution conformers.
Glycobiology, 23, 2013
|
|
1ONI
 
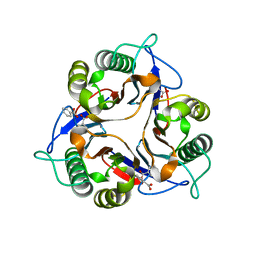 | | Crystal structure of a human p14.5, a translational inhibitor reveals different mode of ligand binding near the invariant residues of the Yjgf/UK114 protein family | | Descriptor: | 14.5 kDa translational inhibitor protein, BENZOIC ACID | | Authors: | Manjasetty, B.A, Delbrueck, H, Mueller, U, Erdmann, M.F, Heinemann, U. | | Deposit date: | 2003-02-28 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Homo sapiens protein hp14.5.
Proteins, 54, 2004
|
|
1XCR
 
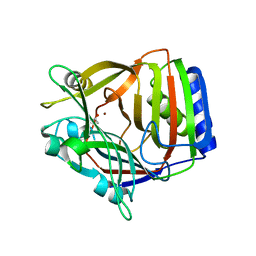 | | Crystal Structure of Longer Splice Variant of PTD012 from Homo sapiens reveals a novel Zinc-containing fold | | Descriptor: | ACETIC ACID, ZINC ION, hypothetical protein PTD012 | | Authors: | Manjasetty, B.A, Fieber-Erdmann, M, Roske, Y, Goetz, F, Buessow, K, Heinemann, U. | | Deposit date: | 2004-09-03 | | Release date: | 2005-09-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Homo sapiens PTD012 reveals a zinc-containing hydrolase fold
Protein Sci., 15, 2006
|
|
7NDJ
 
 | |
4URR
 
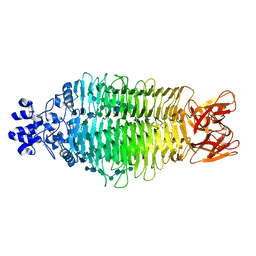 | | Tailspike protein of Sf6 bacteriophage bound to Shigella flexneri O- antigen octasaccharide fragment | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL TAIL PROTEIN, MANGANESE (II) ION, ... | | Authors: | Gohlke, U, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2014-07-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bacteriophage Tailspikes and Bacterial O-Antigens as a Model System to Study Weak-Affinity Protein-Polysaccharide Interactions.
J.Am.Chem.Soc., 138, 2016
|
|
1OQA
 
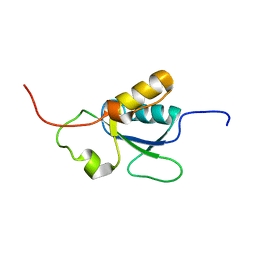 | | Solution structure of the BRCT-c domain from human BRCA1 | | Descriptor: | Breast cancer type 1 susceptibility protein | | Authors: | Gaiser, O.J, Ball, L.J, Schmieder, P, Leitner, D, Strauss, H, Wahl, M, Kuhne, R, Oschkinat, H, Heinemann, U. | | Deposit date: | 2003-03-07 | | Release date: | 2004-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure, backbone dynamics, and association behavior of the C-terminal BRCT domain from the breast cancer-associated protein BRCA1.
Biochemistry, 43, 2004
|
|
