2IVO
 
 | | Structure of UP1 protein | | Descriptor: | TUNGSTATE(VI)ION, UP1 | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
2IVN
 
 | | Structure of UP1 protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, O-SIALOGLYCOPROTEIN ENDOPEPTIDASE, ... | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
2IVP
 
 | | Structure of UP1 protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FE (II) ION, O-SIALOGLYCOPROTEIN ENDOPEPTIDASE | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
2VWB
 
 | | Structure of the archaeal Kae1-Bud32 fusion protein MJ1130: a model for the eukaryotic EKC-KEOPS subcomplex involved in transcription and telomere homeostasis. | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PUTATIVE O-SIALOGLYCOPROTEIN ENDOPEPTIDASE | | Authors: | Hecker, A, Lopreiato, R, Graille, M, Collinet, B, Forterre, P, Domenico, L, van Tilbeurgh, H. | | Deposit date: | 2008-06-20 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of the Archaeal Kae1/Bud32 Fusion Protein Mj1130: A Model for the Eukaryotic Ekc/Keops Subcomplex
Embo J., 27, 2008
|
|
9EO0
 
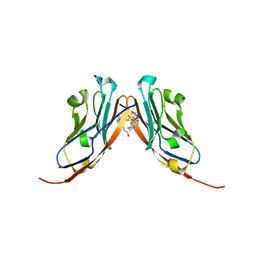 | | Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 | | Descriptor: | Programmed cell death 1 ligand 1, SULFATE ION, ~{N}-[3-[3-[[5-[(2-hydroxyethylamino)methyl]pyridin-2-yl]carbonylamino]-2-methyl-phenyl]-2-methyl-phenyl]-5-[[3-(methylsulfonylamino)propylamino]methyl]pyridine-2-carboxamide | | Authors: | Plewka, J, Hec, A, Sitar, T, Holak, T. | | Deposit date: | 2024-03-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nonsymmetrically Substituted 1,1'-Biphenyl-Based Small Molecule Inhibitors of the PD-1/PD-L1 Interaction.
Acs Med.Chem.Lett., 15, 2024
|
|
4USS
 
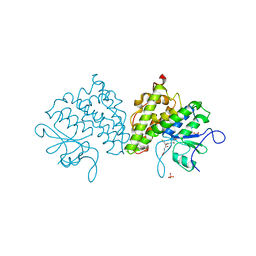 | | Populus trichocarpa glutathione transferase X1-1 (GHR1), complexed with glutathione | | Descriptor: | GLUTATHIONE, GLUTATHIONYL HYDROQUINONE REDUCTASE, PHOSPHATE ION | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Dumaracay, S, Favier, F, Didierjean, C, Saul, F, Haouz, A, Morel-Rouhier, M, Gelhaye, E, Rouhier, N, Hecker, A. | | Deposit date: | 2014-07-13 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutathionyl-Hydroquinone Reductases from Poplar are Plastidial Proteins that Deglutathionylate Both Reduced and Oxidized Glutathionylated Quinones.
FEBS Lett., 589, 2015
|
|
4PQH
 
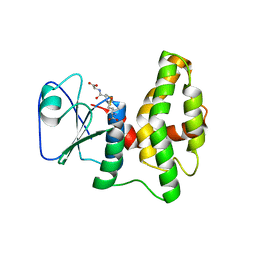 | | Crystal structure of glutathione transferase lambda1 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, SODIUM ION, glutathione transferase lambda1 | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
5HVZ
 
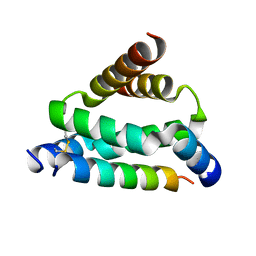 | | Crystal structure of smAKAP AKB domain bound RIa dimerization/docking (D/D) complex at 2.0 A resolution | | Descriptor: | Small membrane A-kinase anchor protein, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Wu, J, Burgers, P.P, Bruystens, J, Heck, A.J.R, Taylor, S.S. | | Deposit date: | 2016-01-28 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of smAKAP and its regulation by PKA-mediated phosphorylation.
Febs J., 283, 2016
|
|
7PKA
 
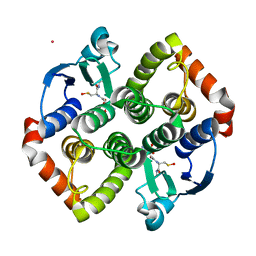 | | Synechocystis sp. PCC6803 glutathione transferase Chi 1, GSOH bound | | Descriptor: | Glutathione S-transferase, POTASSIUM ION, S-Hydroxy-Glutathione | | Authors: | Didierjean, C, Mocchetti, E, Hecker, A, Favier, F. | | Deposit date: | 2021-08-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure and functions of glutathione transferase Chi 1 from cyanobacterium Synechocystis sp. PCC 6803
To Be Published
|
|
6SGO
 
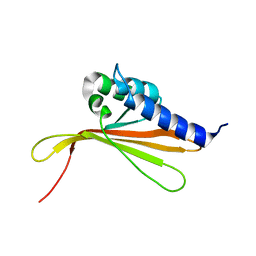 | | NMR structure of MLP124017 | | Descriptor: | Secreted protein | | Authors: | Barthe, P, de Guillen, K, Padilla, A, Hecker, A. | | Deposit date: | 2019-08-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural genomics applied to the rust fungus Melampsora larici-populina reveals two candidate effector proteins adopting cystine knot and NTF2-like protein folds.
Sci Rep, 9, 2019
|
|
6P73
 
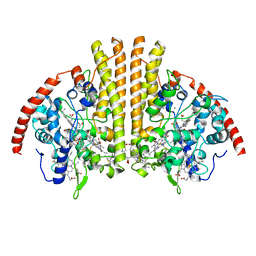 | | Cytochrome-C-nitrite reductase | | Descriptor: | CALCIUM ION, Cytochrome c-552, HEME C | | Authors: | Schmidt, M, Pacheco, A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Trapping of a Putative Intermediate in the CytochromecNitrite Reductase (ccNiR)-Catalyzed Reduction of Nitrite: Implications for the ccNiR Reaction Mechanism.
J.Am.Chem.Soc., 141, 2019
|
|
6H0I
 
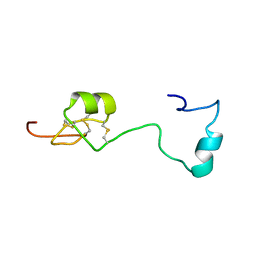 | | Solution structure of Melampsora larici-populina MlpP4.1 | | Descriptor: | Secreted protein | | Authors: | Tsan, P, Petre, B, Hecker, A, Rouhier, N, Duplessis, S. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural genomics applied to the rust fungus Melampsora larici-populina reveals two candidate effector proteins adopting cystine knot and NTF2-like protein folds.
Sci Rep, 9, 2019
|
|
6QLE
 
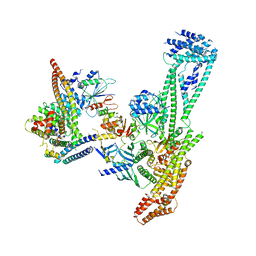 | | Structure of inner kinetochore CCAN complex | | Descriptor: | Central kinetochore subunit CTF3,Inner kinetochore subunit CTF3,Central kinetochore subunit CTF3,Inner kinetochore subunit CTF3, Central kinetochore subunit MCM16,Central kinetochore subunit MCM16,Inner kinetochore subunit MCM16,Mcm16p, Inner kinetochore subunit AME1,Inner kinetochore subunit AME1,Inner kinetochore subunit AME1,Inner kinetochore subunit AME1, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
6QLD
 
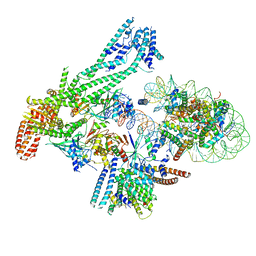 | | Structure of inner kinetochore CCAN-Cenp-A complex | | Descriptor: | DNA (125-MER), Histone H2A.1, Histone H2B.1, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
6QLF
 
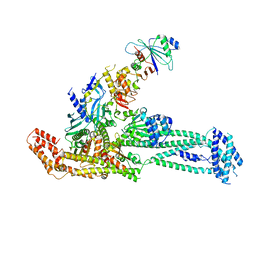 | | Structure of inner kinetochore CCAN complex with mask1 | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CHL4, Inner kinetochore subunit CTF19, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
5DYN
 
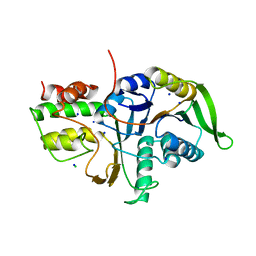 | | B. fragilis cysteine protease | | Descriptor: | CHLORIDE ION, Putative peptidase, SODIUM ION | | Authors: | Choi, V.M, Herrou, J, Hecht, A.L, Turner, J.R, Crosson, S, Bubeck Wardenburg, J. | | Deposit date: | 2015-09-24 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice.
Nat. Med., 22, 2016
|
|
5NBT
 
 | | Apo structure of p60N/p80C katanin | | Descriptor: | Katanin p60 ATPase-containing subunit A1, Katanin p80 WD40 repeat-containing subunit B1 | | Authors: | Jiang, K, Rezabkova, L, Hua, S, Liu, Q, Capitani, G, Altelaar, A.F.M, Heck, A.J.R, Kammerer, R.A, Steinmetz, M.O, Akhmanova, A. | | Deposit date: | 2017-03-02 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex.
Nat. Cell Biol., 19, 2017
|
|
5N8Y
 
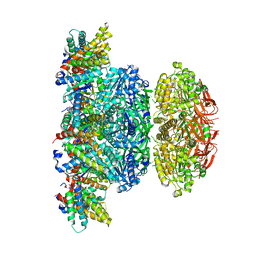 | | KaiCBA circadian clock backbone model based on a Cryo-EM density | | Descriptor: | Circadian clock protein KaiA, Circadian clock protein KaiB, Circadian clock protein kinase KaiC | | Authors: | Schuller, J.M, Snijder, J, Loessl, P, Heck, A.J.R, Foerster, F. | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state.
Science, 355, 2017
|
|
5LKD
 
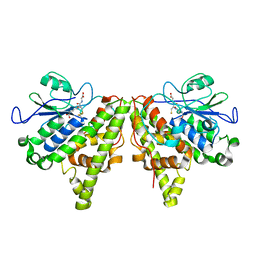 | | Crystal structure of the Xi glutathione transferase ECM4 from Saccharomyces cerevisiae in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase omega-like 2 | | Authors: | Schwartz, M, Didierjean, C, Hecker, A, Girardet, J.M, Morel-Rouhier, M, Gelhaye, E, Favier, F. | | Deposit date: | 2016-07-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae ECM4, a Xi-Class Glutathione Transferase that Reacts with Glutathionyl-(hydro)quinones.
Plos One, 11, 2016
|
|
5LKB
 
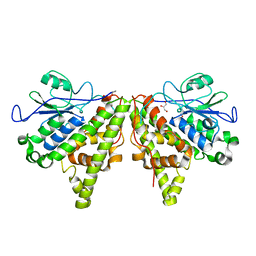 | | Crystal structure of the Xi glutathione transferase ECM4 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Glutathione S-transferase omega-like 2 | | Authors: | Schwartz, M, Didierjean, C, Hecker, A, Girardet, J.M, Morel-Rouhier, M, Gelhaye, E, Favier, F. | | Deposit date: | 2016-07-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Saccharomyces cerevisiae ECM4, a Xi-Class Glutathione Transferase that Reacts with Glutathionyl-(hydro)quinones.
Plos One, 11, 2016
|
|
5NMR
 
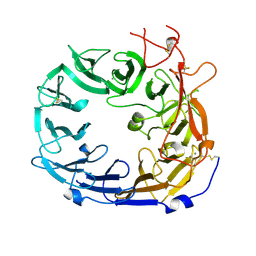 | | Monomeric mouse Sortilin extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Leloup, N.O.L, Loessl, P, Meijer, D.H.M, Heck, A.J.R, Thies-Weesie, D.M.E, Janssen, B.J.C. | | Deposit date: | 2017-04-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Low pH-induced conformational change and dimerization of sortilin triggers endocytosed ligand release.
Nat Commun, 8, 2017
|
|
4PQI
 
 | | Crystal structure of glutathione transferase lambda3 from Populus trichocarpa | | Descriptor: | CALCIUM ION, GLUTATHIONE, In2-1 family protein, ... | | Authors: | Lallement, P.A, Meux, E, Gualberto, J.M, Prosper, P, Didierjean, C, Haouz, A, Saul, F, Rouhier, N, Hecker, A. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and enzymatic insights into Lambda glutathione transferases from Populus trichocarpa, monomeric enzymes constituting an early divergent class specific to terrestrial plants.
Biochem.J., 462, 2014
|
|
3NFI
 
 | | Crystal structure of tandem winged helix domain of RNA polymerase I subunit A49 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DNA-directed RNA polymerase I subunit RPA49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
3NFH
 
 | | Crystal structure of tandem winged helix domain of RNA polymerase I subunit A49 (P4) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
1LQT
 
 | | A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase | | Descriptor: | 4-OXO-NICOTINAMIDE-ADENINE DINUCLEOTIDE PHOSPHATE, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bossi, R.T, Aliverti, A, Raimondi, D, Fischer, F, Zanetti, G, Ferrari, D, Tahallah, N, Maier, C.S, Heck, A.J.R, Rizzi, M, Mattevi, A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-05-13 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A covalent modification of NADP+ revealed by the atomic resolution structure of FprA, a Mycobacterium tuberculosis oxidoreductase.
Biochemistry, 41, 2002
|
|
