8V3X
 
 | |
8V40
 
 | |
7DV4
 
 | | Crystal structure of anti-CTLA-4 VH domain in complex with human CTLA-4 | | Descriptor: | 1,2-ETHANEDIOL, 4003-1(VH), Cytotoxic T-lymphocyte protein 4 | | Authors: | Li, H, Gan, X, He, Y. | | Deposit date: | 2021-01-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | An anti-CTLA-4 heavy chain-only antibody with enhanced T reg depletion shows excellent preclinical efficacy and safety profile.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8W6I
 
 | |
8W6J
 
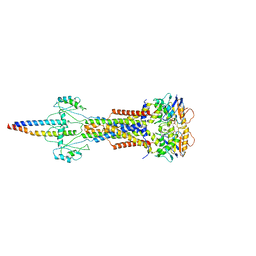 | | Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC complex with ATP in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Li, J, Xu, X, He, Y, Luo, M. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of Escherichia coli Str K12 FtsE(E163Q)X/EnvC complex with ATP in peptidisc
To Be Published
|
|
7DPX
 
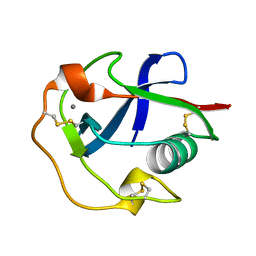 | |
7DVA
 
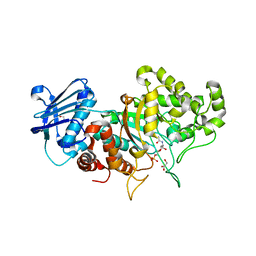 | | Structure of wild type Bt4394, a GH20 family sulfoglycosidase, in complex with 6S-GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DUP
 
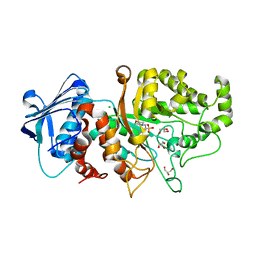 | | Apo structure of wild type Bt4394, a GH20 family sulfoglycosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVB
 
 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
8GA8
 
 | | Structure of the yeast (HDAC) Rpd3L complex | | Descriptor: | Histone deacetylase RPD3, Transcriptional regulatory protein DEP1, Transcriptional regulatory protein PHO23, ... | | Authors: | Patel, A.B, Radhakrishnan, I, He, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the Saccharomyces cerevisiae Rpd3L histone deacetylase complex.
Nat Commun, 14, 2023
|
|
6JLI
 
 | | Crystal structure of CTLD7 domain of human PLA2R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Secretory phospholipase A2 receptor | | Authors: | Yu, B, Hu, Z, Kong, D, Cheng, C, He, Y. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | Crystal structure of the CTLD7 domain of human M-type phospholipase A2 receptor.
J.Struct.Biol., 207, 2019
|
|
7EK6
 
 | | Structure of viral peptides IPB19/N52 | | Descriptor: | Spike protein S2 | | Authors: | Yu, D, Qin, B, Cui, S, He, Y. | | Deposit date: | 2021-04-04 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.243 Å) | | Cite: | Structure-based design and characterization of novel fusion-inhibitory lipopeptides against SARS-CoV-2 and emerging variants.
Emerg Microbes Infect, 10, 2021
|
|
5XBF
 
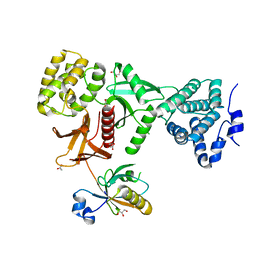 | | Crystal Structure of Myo7b C-terminal MyTH4-FERM in complex with USH1C PDZ3 | | Descriptor: | ACETATE ION, D-MALATE, GLYCEROL, ... | | Authors: | Li, J, He, Y, Weck, W.L, Lu, Q, Tyska, M.J, Zhang, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure of Myo7b/USH1C complex suggests a general PDZ domain binding mode by MyTH4-FERM myosins.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7WHW
 
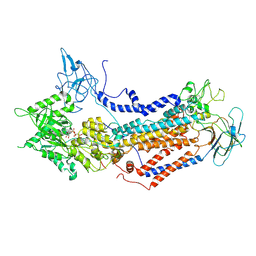 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in detergent with AMPPCP (E1-ATP state) | | Descriptor: | Alkylphosphocholine resistance protein LEM3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7WHV
 
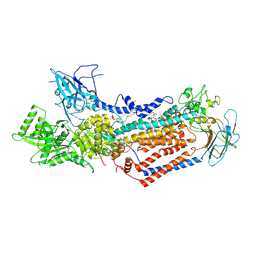 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in detergent with beryllium fluoride (E2P state) | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7F7F
 
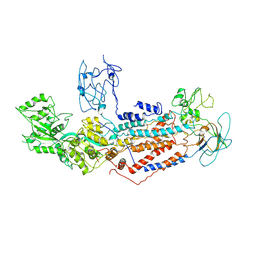 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in yeast lipids with beryllium fluoride (resting state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-06-29 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DRX
 
 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with beryllium fluoride (E2P state) | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-30 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DSH
 
 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with AMPPCP (E1-ATP state) | | Descriptor: | Alkylphosphocholine resistance protein LEM3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DSI
 
 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in yeast lipids with AMPPCP ( resting state ) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
3FXV
 
 | | Identification of an N-oxide pyridine GW4064 analogue as a potent FXR agonist | | Descriptor: | 12-meric peptide from Nuclear receptor coactivator 1, 6-(4-{[3-(3,5-dichloropyridin-4-yl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-2-methylphenyl)-1-methyl-1H-indole-3-carbox ylic acid, NR1H4 protein | | Authors: | Feng, S, Yang, M, He, Y, Chen, L, Zhang, Z, Wang, Z, Hong, D, Richter, H, Benson, G.M, Bleicher, K, Grether, U, Martin, R, Plancher, J.-M, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2009-01-21 | | Release date: | 2009-04-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Identification of an N-oxide pyridine GW4064 analog as a potent FXR agonist
Bioorg.Med.Chem.Lett., 19, 2009
|
|
