5YYC
 
 | | Crystal structure of alanine racemase from Bacillus pseudofirmus (OF4) | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dong, H, Hu, T.T, He, G.Z, Lu, D.R, Qi, J.X, Dou, Y.S, Long, W, He, X, Su, D, Ju, J.S. | | Deposit date: | 2017-12-08 | | Release date: | 2019-01-02 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural features and kinetic characterization of alanine racemase from Bacillus pseudofirmus OF4.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
8K4N
 
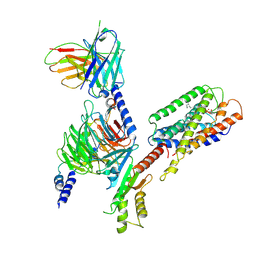 | | Structure of GPR34-Gi complex | | Descriptor: | (2R)-2-azanyl-3-[oxidanyl-[(2R)-2-oxidanyl-3-tetradec-9-enoyloxy-propoxy]phosphoryl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wong, T.S, Zeng, Z.C, Xiong, T.T, Gan, S.Y, He, G.D, Du, Y. | | Deposit date: | 2023-07-20 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM structure of GPR34-Gi complex
To Be Published
|
|
1TWI
 
 | | Crystal structure of Diaminopimelate Decarboxylase from m. jannaschii in co-complex with L-lysine | | Descriptor: | Diaminopimelate decarboxylase, LYSINE, MAGNESIUM ION, ... | | Authors: | Rajashankar, K.R, Ray, S.S, Bonanno, J.B, Pinho, M.G, He, G, De Lencastre, H, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-01 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic resistance accessory factor
Structure, 10, 2002
|
|
1TUF
 
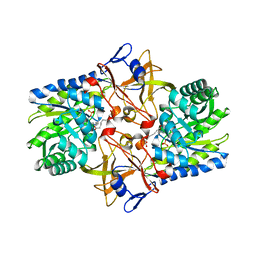 | | Crystal structure of Diaminopimelate Decarboxylase from m. jannaschi | | Descriptor: | AZELAIC ACID, Diaminopimelate decarboxylase | | Authors: | Rajashankar, K, Ray, S.R, Bonanno, J.B, Pinho, M.G, He, G, De Lencastre, H, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-24 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic resistance accessory factor
Structure, 10, 2002
|
|
8JTZ
 
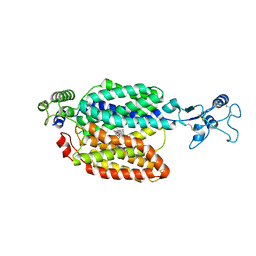 | | hOCT1 in complex with spironolactone in outward facing partially occluded conformation | | Descriptor: | SPIRONOLACTONE, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTX
 
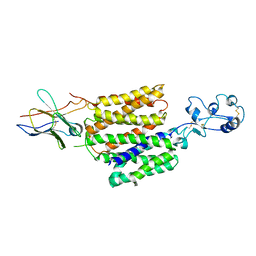 | | hOCT1 in complex with nb5660 in inward facing fully open conformation | | Descriptor: | Solute carrier family 22 member 1, nanobody 56 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JU0
 
 | | hOCT1 in complex with spironolactone in inward facing occluded conformation | | Descriptor: | SPIRONOLACTONE, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-23 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTS
 
 | | hOCT1 in complex with metformin in outward open conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTV
 
 | | hOCT1 in complex with metformin in inward occluded conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTT
 
 | | hOCT1 in complex with metformin in outward occluded conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTW
 
 | | hOCT1 in complex with nb5660 in inward facing partially open 1 conformation | | Descriptor: | Solute carrier family 22 member 1, nanobody 56 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTY
 
 | | hOCT1 in complex with nb5660 in inward facing partially open 2 conformation | | Descriptor: | Solute carrier family 22 member 1, nanobody 56 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
