6HBC
 
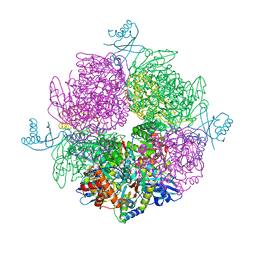 | | Structure of the repeat unit in the network formed by CcmM and Rubisco from Synechococcus elongatus | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, Ribulose 1,5-bisphosphate carboxylase small subunit, Ribulose bisphosphate carboxylase large chain | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6TMX
 
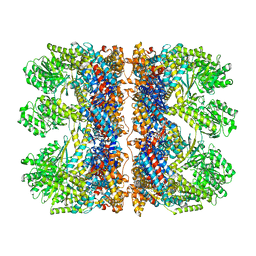 | | Structure of the chaperonin gp146 from the bacteriophage EL (Pseudomonas aeruginosa) in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bracher, A, Wang, H, Paul, S.S, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
6TMU
 
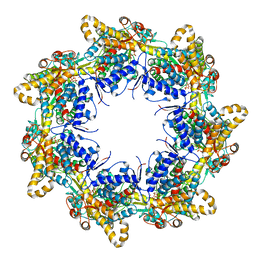 | | Crystal structure of the chaperonin gp146 from the bacteriophage EL 2 (Pseudomonas aeruginosa) in presence of ATP-BeFx, crystal form II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bracher, A, Paul, S.S, Wang, H, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
6HBB
 
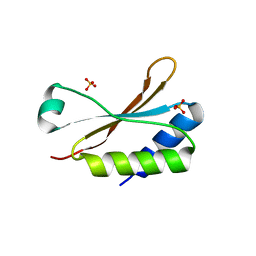 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, SULFATE ION | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6TMW
 
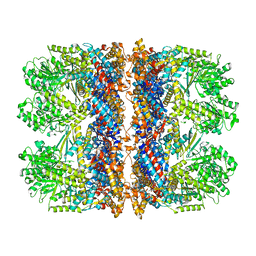 | | Structure of the chaperonin gp146 from the bacteriophage EL (Pseudomonas aeruginosa) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative GroEL-like chaperonine protein | | Authors: | Bracher, A, Wang, H, Paul, S.S, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.91 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
6TMT
 
 | | Crystal structure of the chaperonin gp146 from the bacteriophage EL 2 (Pseudomonas aeruginosa) in presence of ATP-BeFx, crystal form I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative GroEL-like chaperonine protein | | Authors: | Bracher, A, Paul, S.S, Wang, H, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.03 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
6H6G
 
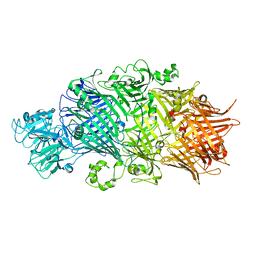 | | Crystal Structure of TcdB2-TccC3 without hypervariable C-terminal region | | Descriptor: | TcdB2,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
6TMV
 
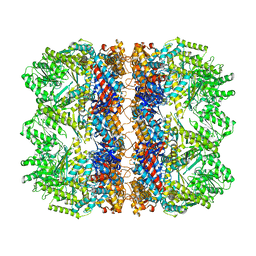 | | Structure of the chaperonin gp146 from the bacteriophage EL (Pseudomonas aeruginosa) in the apo state | | Descriptor: | Putative GroEL-like chaperonine protein | | Authors: | Bracher, A, Wang, H, Paul, S.S, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
3SYK
 
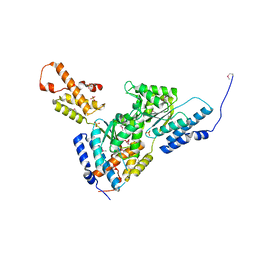 | | Crystal structure of the AAA+ protein CbbX, selenomethionine structure | | Descriptor: | Protein CbbX, SULFATE ION | | Authors: | Mueller-Cajar, O, Stotz, M, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase.
Nature, 479, 2011
|
|
3SYL
 
 | | Crystal structure of the AAA+ protein CbbX, native structure | | Descriptor: | Protein CbbX, SULFATE ION | | Authors: | Mueller-Cajar, O, Stotz, M, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase.
Nature, 479, 2011
|
|
6HZK
 
 | | Crystal structure of redox-inhibited phosphoribulokinase from Synechococcus sp. (strain PCC 6301) | | Descriptor: | Phosphoribulokinase | | Authors: | Wilson, R.H, Bracher, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of phosphoribulokinase from Synechococcus sp. strain PCC 6301.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6HZL
 
 | | Crystal structure of redox-inhibited phosphoribulokinase from Synechococcus sp. (strain PCC 6301), osmate derivative | | Descriptor: | OSMIUM ION, Phosphoribulokinase | | Authors: | Wilson, R.H, Bracher, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-27 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of phosphoribulokinase from Synechococcus sp. strain PCC 6301.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6H6F
 
 | | PTC3 holotoxin complex from Photorhabdus luminiscens - Mutant TcC-D651A | | Descriptor: | TcdA1, TcdB2,TccC3,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
6H6E
 
 | | PTC3 holotoxin complex from Photorhabdus luminecens in prepore state (TcdA1, TcdB2, TccC3) | | Descriptor: | TcdA1, TcdB2,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
3ZUH
 
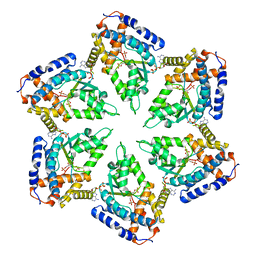 | | Negative stain EM Map of the AAA protein CbbX, a red-type Rubisco activase from R. sphaeroides | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PROTEIN CBBX, RIBULOSE-1,5-DIPHOSPHATE | | Authors: | Mueller-Cajar, O, Stotz, M, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Structure and Function of the Aaa+ Protein Cbbx, a Red-Type Rubisco Activase.
Nature, 479, 2011
|
|
3T15
 
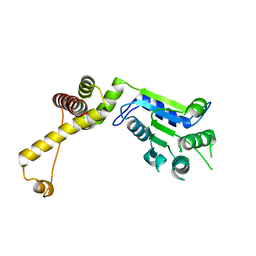 | | Structure of green-type Rubisco activase from tobacco | | Descriptor: | Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplastic | | Authors: | Stotz, M, Wendler, P, Mueller-Cajar, O, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-21 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of green-type Rubisco activase from tobacco.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3ZW6
 
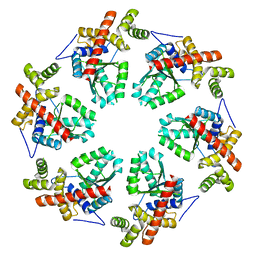 | | MODEL OF HEXAMERIC AAA DOMAIN ARRANGEMENT OF GREEN-TYPE RUBISCO ACTIVASE FROM TOBACCO. | | Descriptor: | RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE ACTIVASE 1, CHLOROPLASTIC | | Authors: | Stotz, M, Mueller-Cajar, O, Ciniawsky, S, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure of Green-Type Rubisco Activase from Tobacco
Nat.Struct.Mol.Biol., 18, 2011
|
|
5D5U
 
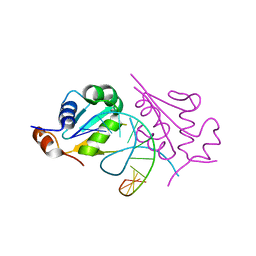 | | Crystal structure of human Hsf1 with HSE DNA | | Descriptor: | Heat shock Element DNA, Heat shock factor protein 1 | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D60
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form III | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5W
 
 | | Crystal structure of Chaetomium thermophilum Skn7 with HSE DNA | | Descriptor: | HSE DNA, Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5V
 
 | | Crystal structure of human Hsf1 with Satellite III repeat DNA | | Descriptor: | DNA, Heat shock factor protein 1, MAGNESIUM ION | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5X
 
 | | Crystal structure of Chaetomium thermophilum Skn7 with SSRE DNA | | Descriptor: | Putative transcription factor, SSRE DNA strand 1, SSRE DNA strand 2 | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5Y
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form I | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5Z
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form II | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Bracher, A, Hartl, F.U. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2PEN
 
 | | Crystal structure of RbcX, crystal form I | | Descriptor: | ORF134 | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
