4RNS
 
 | | PcpR inducer binding domain (apo-form) | | Descriptor: | PCP degradation transcriptional activation protein | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-25 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
4RPO
 
 | | PcpR inducer binding domain (Complex with 2,4,6-trichlorophenol) | | Descriptor: | 2,4,6-trichlorophenol, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
4RPN
 
 | | PcpR inducer binding domain complex with pentachlorophenol | | Descriptor: | PCP degradation transcriptional activation protein, PENTACHLOROPHENOL | | Authors: | Hayes, R.P, Moural, T.W, Lewis, K.M, Onofrei, D, Xun, L, Kang, C. | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Structures of the Inducer-Binding Domain of Pentachlorophenol-Degrading Gene Regulator PcpR from Sphingobium chlorophenolicum.
Int J Mol Sci, 15, 2014
|
|
3S2I
 
 | | Crystal Structure of FurX NADH+:Furfuryl alcohol II | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
5H9F
 
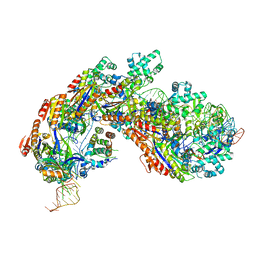 | | Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target at 2.45 angstrom resolution. | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Hayes, R.P, Xiao, Y, Ding, F, van Erp, P.B.G, Rajashankar, K, Bailey, S, Wiedenheft, B, Ke, A. | | Deposit date: | 2015-12-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.
Nature, 530, 2016
|
|
7LUF
 
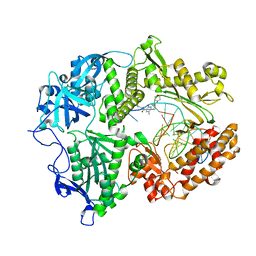 | | HSV1 polymerase ternary complex with dsDNA and PNU-183792 | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*AP*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*CP*G)-3'), DNA polymerase, ... | | Authors: | Hayes, R.P, Klein, D. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural understanding of non-nucleoside inhibition in an elongating herpesvirus polymerase.
Nat Commun, 12, 2021
|
|
8DI4
 
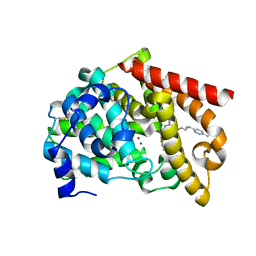 | | Discovery of MK-8189, a highly potent and selective PDE10A inhibitor for the treatment of schizophrenia | | Descriptor: | 2-methyl-6-{[(1S,2S)-2-(5-methylpyridin-2-yl)cyclopropyl]methoxy}-N-[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]pyrimidin-4-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Hayes, R.P, Yan, Y. | | Deposit date: | 2022-06-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Discovery of MK-8189, a Highly Potent and Selective PDE10A Inhibitor for the Treatment of Schizophrenia.
J.Med.Chem., 66, 2023
|
|
5H9E
 
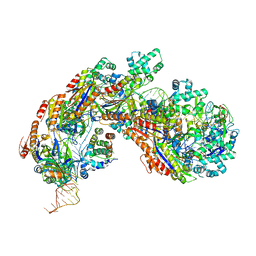 | | Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target (32-nt spacer) at 3.20 angstrom resolution. | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Hayes, R.P, Xiao, Y, Ding, F, van Erp, P.B.G, Rajashankar, K, Bailey, S, Wiedenheft, B, Ke, A. | | Deposit date: | 2015-12-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli.
Nature, 530, 2016
|
|
4HUZ
 
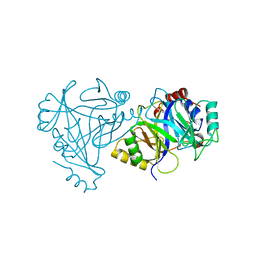 | | 2,6-Dichloro-p-hydroquinone 1,2-Dioxygenase | | Descriptor: | 2,6-dichloro-p-hydroquinone 1,2-dioxygenase, FE (III) ION, SULFATE ION | | Authors: | Hayes, R.P, Nissen, M.S, Green, A.R, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2012-11-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of 2,6-dichloro-p-hydroquinone 1,2-dioxygenase (PcpA) from Sphingobium chlorophenolicum, a new type of aromatic ring-cleavage enzyme.
Mol.Microbiol., 88, 2013
|
|
4LBI
 
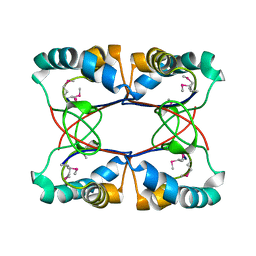 | |
4LBH
 
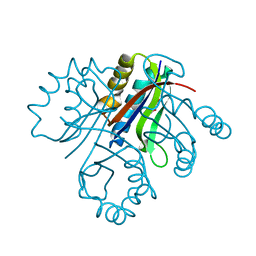 | | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) from Burkholderia phenoliruptrix AC1100: Apo-form | | Descriptor: | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG), DI(HYDROXYETHYL)ETHER | | Authors: | Hayes, R.P, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalytic Mechanism of 5-Chlorohydroxyhydroquinone Dehydrochlorinase from the YCII Superfamily of Largely Unknown Function.
J.Biol.Chem., 288, 2013
|
|
4LBP
 
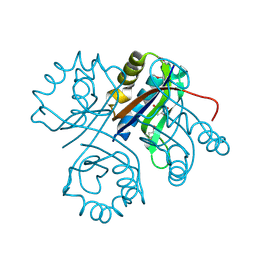 | | 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) from Burkholderia phenoliruptrix AC1100: Complex with 2,5-dihydroxybenzoquinone | | Descriptor: | 2,5-dihydroxycyclohexa-2,5-diene-1,4-dione, 5-chloro-2-hydroxyhydroquinone dehydrochlorinase (TftG) | | Authors: | Hayes, R.P, Lewis, K.M, Xun, L, Kang, C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-28 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Catalytic Mechanism of 5-Chlorohydroxyhydroquinone Dehydrochlorinase from the YCII Superfamily of Largely Unknown Function.
J.Biol.Chem., 288, 2013
|
|
3S2G
 
 | | Crystal Structure of FurX NADH+:Furfuryl alcohol I | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
3S1L
 
 | | Crystal Structure of Apo-form FurX | | Descriptor: | HEXAETHYLENE GLYCOL, ZINC ION, Zinc-containing alcohol dehydrogenase superfamily | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-15 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Furfural reduction mechanism of a zinc-dependent alcohol dehydrogenase from Cupriavidus necator JMP134.
Mol.Microbiol., 83, 2012
|
|
3S2E
 
 | | Crystal Structure of FurX NADH Complex 1 | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal Structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
3S2F
 
 | | Crystal Structure of FurX NADH:Furfural | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHORYLISOPROPANE, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
4G5E
 
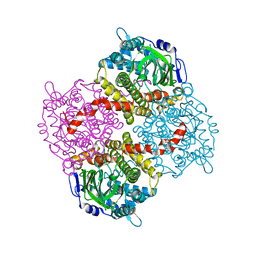 | | 2,4,6-Trichlorophenol 4-monooxygenase | | Descriptor: | 2,4,6-Trichlorophenol 4-monooxygenase | | Authors: | Hayes, R.P, Webb, B.N, Subramanian, A.K, Nissen, M, Popchock, A, Xun, L, Kang, C. | | Deposit date: | 2012-07-17 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Catalytic Differences between Two FADH(2)-Dependent Monooxygenases: 2,4,5-TCP 4-Monooxygenase (TftD) from Burkholderia cepacia AC1100 and 2,4,6-TCP 4-Monooxygenase (TcpA) from Cupriavidus necator JMP134.
Int J Mol Sci, 13, 2012
|
|
6Y7C
 
 | | Early cytoplasmic yeast pre-40S particle (purified with Tsr1 as bait) | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Plassart, L, Plisson-Chastang, C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Good Vibrations: Structural Remodeling of Maturing Yeast Pre-40S Ribosomal Particles Followed by Cryo-Electron Microscopy.
Molecules, 25, 2020
|
|
6OLA
 
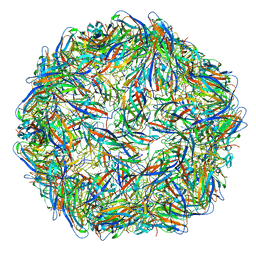 | | Structure of the PCV2d virus-like particle | | Descriptor: | Capsid protein, DNA (5'-D(P*CP*CP*GP*G)-3') | | Authors: | Khayat, R, Wen, K, Alimova, A, Galarza, J, Gottlieb, P. | | Deposit date: | 2019-04-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural characterization of the PCV2d virus-like particle at 3.3 angstrom resolution reveals differences to PCV2a and PCV2b capsids, a tetranucleotide, and an N-terminus near the icosahedral 3-fold axes.
Virology, 537, 2019
|
|
1NJT
 
 | | COMPLEX STRUCTURE OF HCMV PROTEASE AND A PEPTIDOMIMETIC INHIBITOR | | Descriptor: | CHLORIDE ION, Capsid protein P40, Peptidomimetic Inhibitor | | Authors: | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
6RBD
 
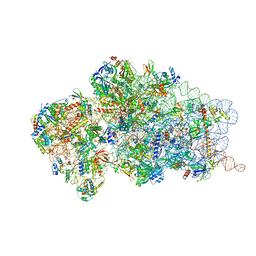 | | State 1 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
6RBE
 
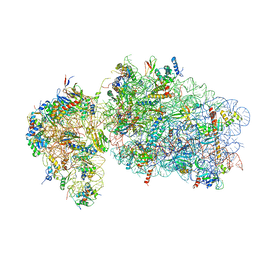 | | State 2 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Shayan, R, Mitterer, V, Ferreira-Cerca, S, Murat, G, Enne, T, Rinaldi, D, Weigl, S, Omanic, H, Gleizes, P.E, Kressler, D, Pertschy, B, Plisson-Chastang, C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism.
Nat Commun, 10, 2019
|
|
1NJU
 
 | | Complex structure of HCMV Protease and a peptidomimetic inhibitor | | Descriptor: | Assemblin, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | Authors: | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
1NKM
 
 | | Complex structure of HCMV Protease and a peptidomimetic inhibitor | | Descriptor: | Assemblin, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | Authors: | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | Deposit date: | 2003-01-03 | | Release date: | 2003-02-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
1NKK
 
 | | COMPLEX STRUCTURE OF HCMV PROTEASE AND A PEPTIDOMIMETIC INHIBITOR | | Descriptor: | Capsid protein P40, Peptidomimetic inhibitor | | Authors: | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | Deposit date: | 2003-01-03 | | Release date: | 2003-02-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
