3CSP
 
 | | Crystal structure of the DM2 mutant of myelin oligodendrocyte glycoprotein | | Descriptor: | Myelin-oligodendrocyte glycoprotein | | Authors: | Breithaupt, C, Schafer, B, Pellkofer, H, Huber, R, Linington, C, Jacob, U. | | Deposit date: | 2008-04-10 | | Release date: | 2008-10-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demyelinating myelin oligodendrocyte glycoprotein-specific autoantibody response is focused on one dominant conformational epitope region in rodents
J.Immunol., 181, 2008
|
|
2HSA
 
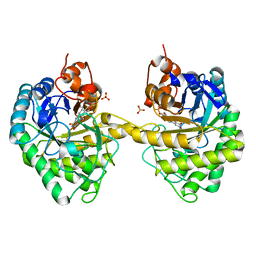 | | Crystal structure of 12-oxophytodienoate reductase 3 (OPR3) from tomato | | Descriptor: | 12-oxophytodienoate reductase 3, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Breithaupt, C, Clausen, T, Huber, R. | | Deposit date: | 2006-07-21 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 12-oxophytodienoate reductase 3 from tomato: Self-inhibition by dimerization.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HS6
 
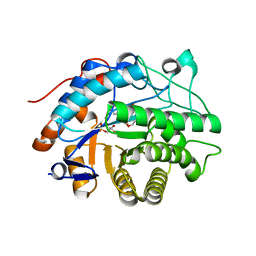 | |
2HS8
 
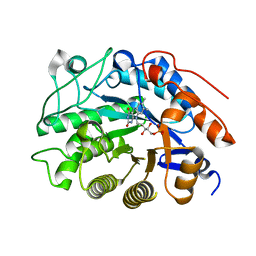 | |
1PKO
 
 | | Myelin Oligodendrocyte Glycoprotein (MOG) | | Descriptor: | Myelin Oligodendrocyte Glycoprotein | | Authors: | Breithaupt, C, Schubart, A, Zander, H, Skerra, A, Huber, R, Linington, C, Jacob, U. | | Deposit date: | 2003-06-06 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the antigenicity of myelin oligodendrocyte glycoprotein
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1ICP
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH PEG400 | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1PKQ
 
 | | Myelin Oligodendrocyte Glycoprotein-(8-18C5) Fab-complex | | Descriptor: | (8-18C5) chimeric Fab, heavy chain, light chain, ... | | Authors: | Breithaupt, C, Schubart, A, Zander, H, Skerra, A, Huber, R, Linington, C, Jacob, U. | | Deposit date: | 2003-06-06 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the antigenicity of myelin oligodendrocyte glycoprotein
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1ICQ
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO COMPLEXED WITH 9R,13R-OPDA | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, 9R,13R-12-OXOPHYTODIENOIC ACID, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
1ICS
 
 | | CRYSTAL STRUCTURE OF 12-OXOPHYTODIENOATE REDUCTASE 1 FROM TOMATO | | Descriptor: | 12-OXOPHYTODIENOATE REDUCTASE 1, FLAVIN MONONUCLEOTIDE | | Authors: | Breithaupt, C, Strassner, J, Breitinger, U, Huber, R, Macheroux, P, Schaller, A, Clausen, T. | | Deposit date: | 2001-04-02 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of 12-oxophytodienoate reductase 1 provides structural insight into substrate binding and specificity within the family of OYE.
Structure, 9, 2001
|
|
5L6Q
 
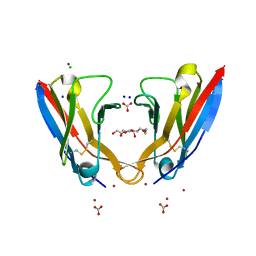 | | Refolded AL protein from cardiac amyloidosis | | Descriptor: | CARBONATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Annamalai, K, Liberta, F, Vielberg, M.-T, Lilie, H, Guehrs, K.-H, Schierhorn, A, Koehler, R, Schmidt, A, Haupt, C, Hegenbart, O, Schoenland, S, Groll, M, Faendrich, M. | | Deposit date: | 2016-05-31 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Common Fibril Structures Imply Systemically Conserved Protein Misfolding Pathways In Vivo.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2AQJ
 
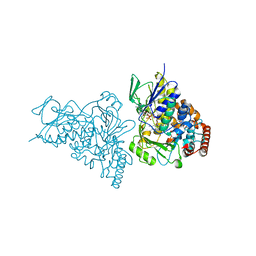 | | The structure of tryptophan 7-halogenase (PrnA) suggests a mechanism for regioselective chlorination | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, TRYPTOPHAN, ... | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.-H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-18 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
2AR8
 
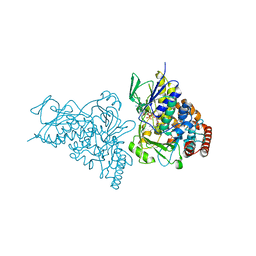 | | The structure of tryptophan 7-halogenase (PrnA)suggests a mechanism for regioselective chlorination | | Descriptor: | 7-CHLOROTRYPTOPHAN, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
2ARD
 
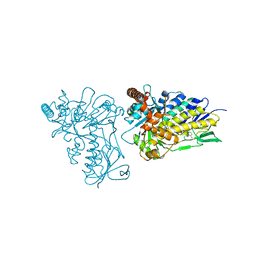 | | The structure of tryptophan 7-halogenase (PrnA) suggests a mechanism for regioselective chlorination | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, tryptophan halogenase PrnA | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
2APG
 
 | | The structure of tryptophan 7-halogenase (PrnA)suggests a mechanism for regioselective chlorination | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-16 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
2A58
 
 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound riboflavin | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION, RIBOFLAVIN | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
2A57
 
 | | Structure of 6,7-Dimthyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 6-carboxyethyl-7-oxo-8-ribityllumazine | | Descriptor: | 3-[8-((2S,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL)-2,4,7-TRIOXO-1,3,8-TRIHYDROPTERIDIN-6-YL]PROPANOIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
2A59
 
 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 5-nitroso-6-ribitylamino-2,4(1H,3H)-pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
7YWC
 
 | | Enzyme of biosynthetic pathway | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, Chorismate dehydratase, GLYCEROL | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2022-02-12 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis.
J.Biol.Chem., 298, 2022
|
|
3HGS
 
 | | Crystal structure of tomato OPR3 in complex with pHB | | Descriptor: | 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Clausen, T, Breithaupt, C. | | Deposit date: | 2009-05-14 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate specificity of plant 12-oxophytodienoate reductases.
J.Mol.Biol., 392, 2009
|
|
3HGO
 
 | |
3HGR
 
 | | Crystal structure of tomato OPR1 in complex with pHB | | Descriptor: | 12-oxophytodienoate reductase 1, FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Clausen, T, Breithaupt, C. | | Deposit date: | 2009-05-14 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of substrate specificity of plant 12-oxophytodienoate reductases.
J.Mol.Biol., 392, 2009
|
|
4E1B
 
 | | Re-refinement of PDB entry 2EQA - SUA5 protein from Sulfolobus tokodaii with bound threonylcarbamoyladenylate | | Descriptor: | MAGNESIUM ION, YrdC/Sua5 family protein, threonylcarbamoyladenylate | | Authors: | Parthier, C, Goerlich, S, Jaenecke, F, Breithaupt, C, Braeuer, U, Fandrich, U, Clausnitzer, D, Wehmeier, U.F, Boettcher, C, Scheel, D, Stubbs, M.T. | | Deposit date: | 2012-03-06 | | Release date: | 2012-03-14 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1SEZ
 
 | | Crystal Structure of Protoporphyrinogen IX Oxidase | | Descriptor: | 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, 4-BROMO-3-(5'-CARBOXY-4'-CHLORO-2'-FLUOROPHENYL)-1-METHYL-5-TRIFLUOROMETHYL-PYRAZOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Koch, M, Breithaupt, C, Kiefersauer, R, Freigang, J, Huber, R, Messerschmidt, A. | | Deposit date: | 2004-02-19 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of protoporphyrinogen IX oxidase: a key enzyme in haem and chlorophyll biosynthesis.
Embo J., 23, 2004
|
|
7AHR
 
 | | Enzyme of biosynthetic pathway | | Descriptor: | 3-[(1-Carboxyvinyl)oxy]benzoic acid, Chorismate dehydratase, GLYCEROL | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2020-09-25 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
7AN5
 
 | | Enzyme of biosynthetic pathway | | Descriptor: | 3-[(1-Carboxyvinyl)oxy]benzoic acid, Chorismate dehydratase | | Authors: | Archna, A, Breithaupt, C, Stubbs, M.T. | | Deposit date: | 2020-10-11 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mechanism of chorismate dehydratase MqnA, the first enzyme of the futalosine pathway, proceeds via substrate-assisted catalysis
J.Biol.Chem., 2022
|
|
