5OCB
 
 | | Crystal structure of nitric oxide bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5OCF
 
 | | Crystal structure of nitric oxide bound to three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5OBO
 
 | | Crystal structure of nitrite bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, GLYCEROL, HEME C, ... | | Authors: | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-06-28 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
1NDT
 
 | | NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (NITRITE REDUCTASE) | | Authors: | Dodd, F.E, Vanbeeumen, J, Eady, R.R, Hasnain, S.S. | | Deposit date: | 1998-10-28 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of a blue-copper nitrite reductase in two crystal forms. The nature of the copper sites, mode of substrate binding and recognition by redox partner.
J.Mol.Biol., 282, 1998
|
|
6Z4K
 
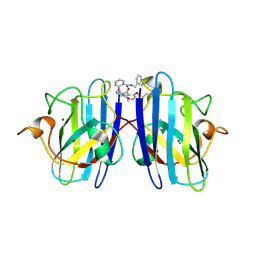 | |
6THF
 
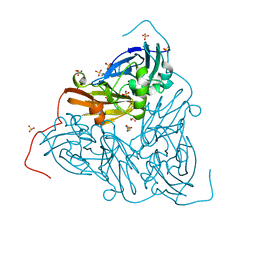 | | Crystal structure of two-domain Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
1E30
 
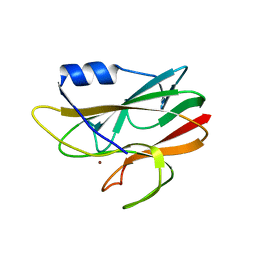 | |
6Z4J
 
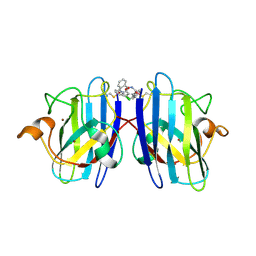 | |
2C9U
 
 | | 1.24 Angstroms resolution structure of as-isolated Cu-Zn Human Superoxide dismutase | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine S, J.S, Hasnain, S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
2C9S
 
 | | 1.24 Angstroms resolution structure of Zn-Zn Human Superoxide dismutase | | Descriptor: | ACETATE ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine S, J.S, Hasnain, S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
6TFD
 
 | | Crystal structure of nitrite and NO bound three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
6FWF
 
 | | Low resolution structure of Neisseria meningitidis qNOR | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | Authors: | Young, D, Antonyuk, S, Tosha, T, Hisano, T, Hasnain, S, Shiro, Y. | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Characterization of the quinol-dependent nitric oxide reductase from the pathogen Neisseria meningitidis, an electrogenic enzyme.
Sci Rep, 8, 2018
|
|
6THE
 
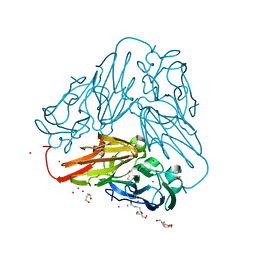 | | Crystal structure of core domain of four-domain heme-cupredoxin-Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6TFO
 
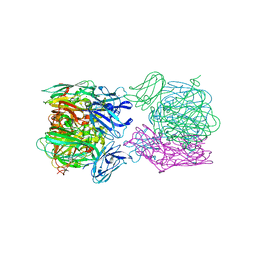 | | Crystal structure of as isolated three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
1GS7
 
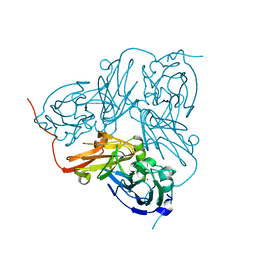 | | Crystal structure of H254F mutant of Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
1GS6
 
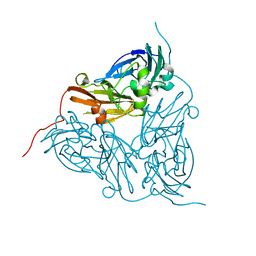 | | Crystal structure of M144A mutant of Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, MAGNESIUM ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
1GS8
 
 | | Crystal structure of mutant D92N Alcaligenes xylosoxidans Nitrite Reductase | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ZINC ION | | Authors: | Ellis, M.J, Prudencio, M, Dodd, F.E, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Crystallographic Studies of the met144Ala, Asp92Asn and His254Phe Mutants of the Nitrite Reductase from Alcaligenes Xylosoxidans Provide Insight Into the Enzyme Mechanism.
J.Mol.Biol., 316, 2002
|
|
6QQ6
 
 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) Val495Ala mutant from Alcaligenes xylosoxidans | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
1E9Q
 
 | | Crystal structure of bovine Cu Zn SOD - (1 of 3) | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hough, M.A, Hasnain, S.S. | | Deposit date: | 2000-10-26 | | Release date: | 2000-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Variability of the Cu Site in One Subunit of Bovine Cuzn Superoxide Dismutase: The Importance of Mobility in the Glu119-Leu142 Loop Region for Catalytic Function
J.Mol.Biol., 304, 2000
|
|
8QGF
 
 | |
1NDR
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF A BLUE COPPER NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Dodd, F.E, Hasnain, S.S, Abraham, Z.H.L, Eady, R.R, Smith, B.E. | | Deposit date: | 1997-01-23 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of a blue-copper nitrite reductase and its substrate-bound complex.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1NDS
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF A SUBSTRATE BOUND BLUE COPPER NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | COPPER (II) ION, NITRITE ION, NITRITE REDUCTASE | | Authors: | Dodd, F.E, Hasnain, S.S, Abraham, Z.H.L, Eady, R.R, Smith, B.E. | | Deposit date: | 1997-01-23 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of a blue-copper nitrite reductase and its substrate-bound complex.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
6T6V
 
 | | Glu-494-Ala inactive monomer of a quinol dependent Nitric Oxide Reductase (qNOR) from Alcaligenes xylosoxidans | | Descriptor: | CALCIUM ION, Nitric oxide reductase subunit B, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gopalasingam, C.C, Johnson, R.M, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-10-19 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
5NMI
 
 | | Cytochrome bc1 bound to the inhibitor MJM170 | | Descriptor: | (4aS)-2-methyl-3-(4-phenoxyphenyl)-5,6,7,8-tetrahydroquinolin-4(4aH)-one, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ARG-ASN-TRP-VAL-PRO-THR-ALA-GLN-LEU-TRP-GLY-ALA-VAL-GLY-ALA-VAL-GLY-LEU-VAL-SER-ALA-THR, ... | | Authors: | Capper, N.J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-04-05 | | Release date: | 2017-06-14 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | New paradigms for understanding and step changes in treating active and chronic, persistent apicomplexan infections.
Sci Rep, 6, 2016
|
|
6Z4M
 
 | |
