7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
5B2C
 
 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, SULFATE ION | | Authors: | Kubota, M, Takeuchi, K, Watanabe, S, Ohno, S, Matsuoka, R, Kohda, D, Hiramatsu, H, Suzuki, Y, Nakayama, T, Terada, T, Shimizu, K, Shimizu, N, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Trisaccharide containing alpha 2,3-linked sialic acid is a receptor for mumps virus
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5B2D
 
 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to 3-sialyllactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kubota, M, Takeuchi, K, Watanabe, S, Ohno, S, Matsuoka, R, Kohda, D, Hiramatsu, H, Suzuki, Y, Nakayama, T, Terada, T, Shimizu, K, Shimizu, N, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Trisaccharide containing alpha 2,3-linked sialic acid is a receptor for mumps virus
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7X7O
 
 | | SARS-CoV-2 spike RBD in complex with neutralizing antibody UT28K | | Descriptor: | Spike protein S1, UT28K Fab, heavy chain, ... | | Authors: | Ozawa, T, Tani, H, Anraku, Y, Kita, S, Igarashi, E, Saga, Y, Inasaki, N, Kawasuji, H, Yamada, H, Sasaki, S, Somekawa, M, Sasaki, J, Hayakawa, Y, Yamamoto, Y, Morinaga, Y, Kurosawa, N, Isobe, M, Fukuhara, H, Maenaka, K, Hashiguchi, T, Kishi, H, Kitajima, I, Saito, S, Niimi, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Novel super-neutralizing antibody UT28K is capable of protecting against infection from a wide variety of SARS-CoV-2 variants.
Mabs, 14, 2022
|
|
6JJN
 
 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to sialyl lewisX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, ... | | Authors: | Kubota, M, Matsuoka, R, Suzuki, T, Yonekura, K, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2019-02-26 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism of the Flexible Glycan Receptor Recognition by Mumps Virus.
J.Virol., 93, 2019
|
|
6JJM
 
 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to the oligosaccharide portion of the GM2 ganglioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, ... | | Authors: | Kubota, M, Matsuoka, R, Suzuki, T, Yonekura, K, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2019-02-26 | | Release date: | 2019-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Molecular Mechanism of the Flexible Glycan Receptor Recognition by Mumps Virus.
J.Virol., 93, 2019
|
|
8IF2
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BQ.1.1 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Kimura, K, Suzuki, T, Hashiguchi, T. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant.
Nat Commun, 14, 2023
|
|
8WXL
 
 | | Structure of the SARS-CoV-2 BA.2.86 spike glycoprotein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-10-30 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XUX
 
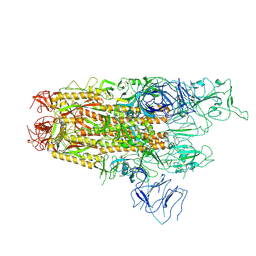 | | Structure of the SARS-CoV-2 BA.2.86 spike protein (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XUZ
 
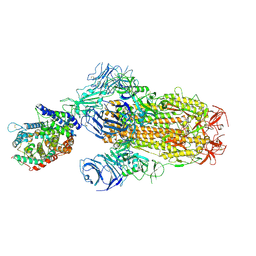 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (2-up and 1-down state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XV0
 
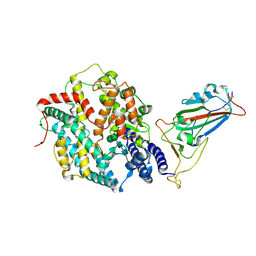 | | Structure of SARS-CoV-2 BA.2.86 spike RBD in complex with ACE2 (up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8XUY
 
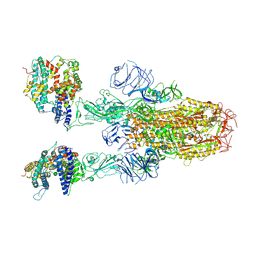 | | Structure of SARS-CoV-2 BA.2.86 spike glycoprotein in complex with ACE2 (2-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2024-01-14 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for receptor-binding domain mobility of the spike in SARS-CoV-2 BA.2.86 and JN.1.
Nat Commun, 15, 2024
|
|
8IOS
 
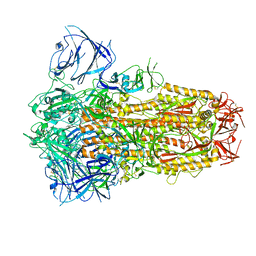 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-1 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOV
 
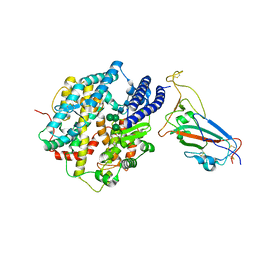 | | Structure of SARS-CoV-2 XBB.1 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOT
 
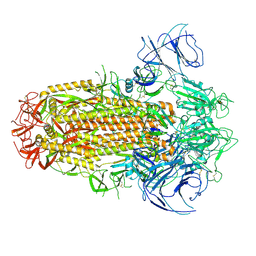 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-2 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOU
 
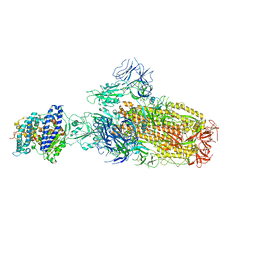 | | Structure of SARS-CoV-2 XBB.1 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IB1
 
 | |
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | Authors: | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
