6XCJ
 
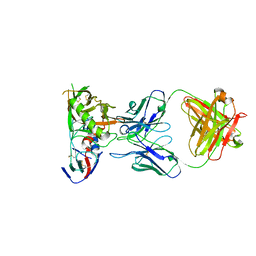 | | Crystal Structure of DH650 Fab from a Rhesus Macaque in Complex with HIV-1 gp120 Core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH650 Fab Heavy Chain, DH650 Fab Light Chain, ... | | Authors: | Raymond, D.D, Chug, H, Harrison, S.C. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recapitulation of HIV-1 Env-antibody coevolution in macaques leading to neutralization breadth.
Science, 371, 2021
|
|
7UI0
 
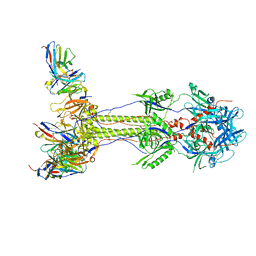 | | Post-fusion ectodomain of HSV-1 gB in complex with HSV010-13 Fab | | Descriptor: | Envelope glycoprotein B, HSV10-13 Fab Heavy chain, HSV10-13 Light chain | | Authors: | Windsor, I.W, Kong, S.L, Garforth, S.J, Almo, S.C, Harrison, S.C. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
J.Clin.Invest., 133, 2023
|
|
7UHZ
 
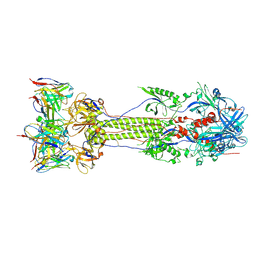 | | Post-fusion ectodomain of HSV-1 gB in complex with BMPC-23 Fab | | Descriptor: | BMPC-23 Fab Heavy chain, BMPC-23 Fab Light chain, Envelope glycoprotein B | | Authors: | Windsor, I.W, Kong, S.L, Garforth, S.J, Almo, S.C, Harrison, S.C. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
J.Clin.Invest., 133, 2023
|
|
6UPH
 
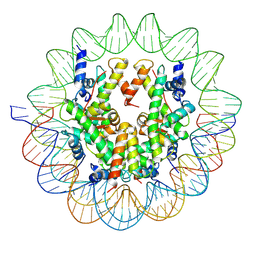 | | Structure of a Yeast Centromeric Nucleosome at 2.7 Angstrom resolution | | Descriptor: | DNA (119-MER), Histone H2A, Histone H2B.1, ... | | Authors: | Migl, D, Kschonsak, M, Arthur, C.P, Khin, Y, Harrison, S.C, Ciferri, C, Dimitrova, Y.N. | | Deposit date: | 2019-10-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryoelectron Microscopy Structure of a Yeast Centromeric Nucleosome at 2.7 angstrom Resolution.
Structure, 28, 2020
|
|
6V05
 
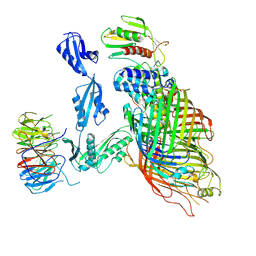 | | Cryo-EM structure of a substrate-engaged Bam complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Tomasek, D, Rawson, S, Lee, J, Wzorek, J.S, Harrison, S.C, Li, Z, Kahne, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a nascent membrane protein as it folds on the BAM complex.
Nature, 583, 2020
|
|
6WXF
 
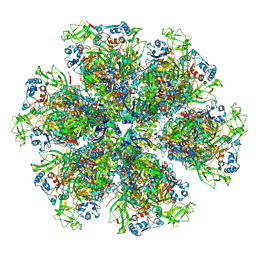 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Intermediate conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
6WXG
 
 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Reversed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
6WXE
 
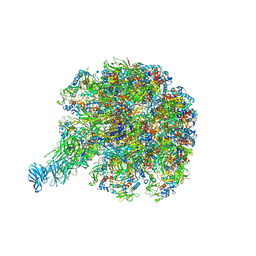 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Upright conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
6WUC
 
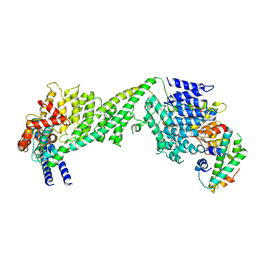 | | The yeast Ctf3 complex with Cnn1-Wip1 | | Descriptor: | Inner kinetochore subunit CNN1, Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, ... | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2020-05-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | The Structural Basis for Kinetochore Stabilization by Cnn1/CENP-T.
Curr.Biol., 30, 2020
|
|
5T58
 
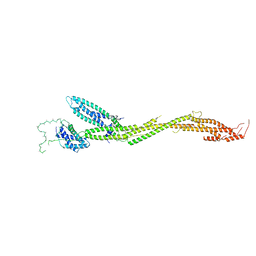 | | Structure of the MIND Complex Shows a Regulatory Focus of Yeast Kinetochore Assembly | | Descriptor: | KLLA0C15939p, KLLA0D15741p, KLLA0E05809p, ... | | Authors: | Dimitrova, Y, Jenni, S, Valverde, R, Khin, Y, Harrison, S.C. | | Deposit date: | 2016-08-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2131 Å) | | Cite: | Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly.
Cell, 167, 2016
|
|
4IT3
 
 | | Crystal Structure of Iml3 from S. cerevisiae | | Descriptor: | Central kinetochore subunit IML3 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2013-01-17 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | An iml3-chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation.
Cell Rep, 5, 2013
|
|
4JE3
 
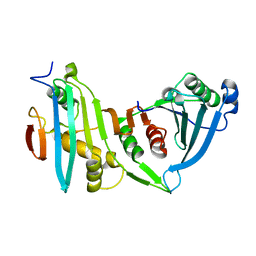 | |
1VPN
 
 | | UNASSEMBLED POLYOMAVIRUS VP1 PENTAMER | | Descriptor: | POLYOMAVIRUS VP1 PENTAMER | | Authors: | Stehle, T, Harrison, S.C. | | Deposit date: | 1997-03-07 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding.
Embo J., 16, 1997
|
|
1VPS
 
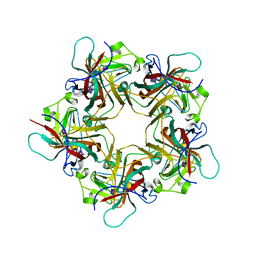 | | POLYOMAVIRUS VP1 PENTAMER COMPLEXED WITH A DISIALYLATED HEXASACCHARIDE | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, POLYOMAVIRUS VP1 PENTAMER | | Authors: | Stehle, T, Harrison, S.C. | | Deposit date: | 1997-03-07 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding.
Embo J., 16, 1997
|
|
1PER
 
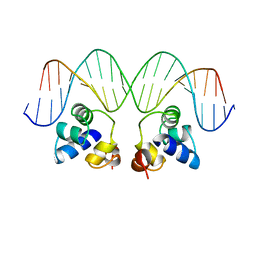 | |
5EN2
 
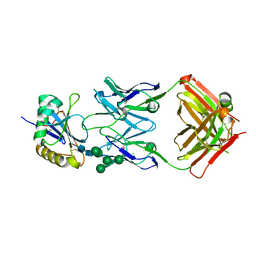 | | Molecular basis for antibody-mediated neutralization of New World hemorrhagic fever mammarenaviruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Mahmutovic, S, Clark, L, Levis, S, Briggiler, A, Enria, D, Harrison, S.C, Abraham, J. | | Deposit date: | 2015-11-09 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Molecular Basis for Antibody-Mediated Neutralization of New World Hemorrhagic Fever Mammarenaviruses.
Cell Host Microbe, 18, 2015
|
|
6OUA
 
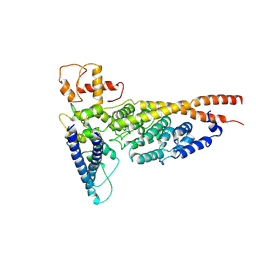 | | Cryo-EM structure of the yeast Ctf3 complex | | Descriptor: | Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, Inner kinetochore subunit MCM22 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2019-05-04 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | The structure of the yeast Ctf3 complex.
Elife, 8, 2019
|
|
1SID
 
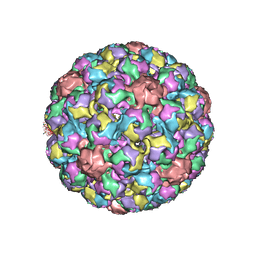 | | MURINE POLYOMAVIRUS COMPLEXED WITH 3'SIALYL LACTOSE | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, POLYOMAVIRUS COAT PROTEIN VP1 | | Authors: | Stehle, T, Harrison, S.C. | | Deposit date: | 1995-12-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments.
Structure, 4, 1996
|
|
5IBU
 
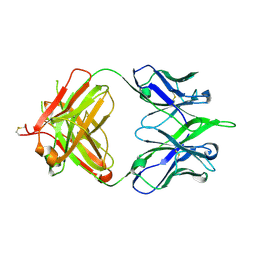 | | 6652 Fab (unbound) | | Descriptor: | 6652 Heavy Chain, 6652 Light Chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2016-02-22 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain.
Nat. Med., 22, 2016
|
|
5IBT
 
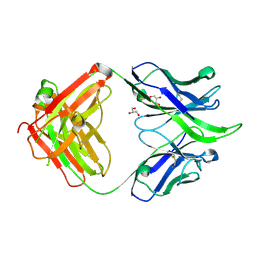 | | UCA Fab (unbound) from 6515 Lineage | | Descriptor: | GLYCEROL, UCA Heavy Chain, UCA Light Chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2016-02-22 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain.
Nat. Med., 22, 2016
|
|
5IBL
 
 | |
4AU6
 
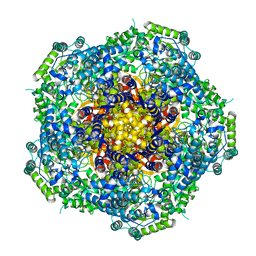 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Location of the Dsrna-Dependent Polymerase, Vp1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
4YJZ
 
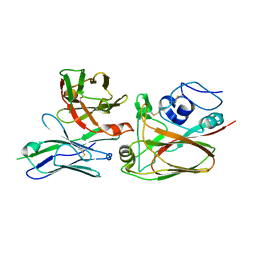 | |
7KQH
 
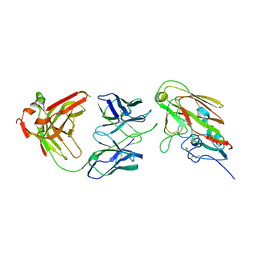 | |
7KQG
 
 | |
