4F5X
 
 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | 分子名称: | Intermediate capsid protein VP6, RNA-directed RNA polymerase, VP2 protein, ... | | 著者 | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | 登録日 | 2012-05-13 | | 公開日 | 2012-10-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (5 Å) | | 主引用文献 | Location of the dsRNA-Dependent Polymerase, VP1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
1RH5
 
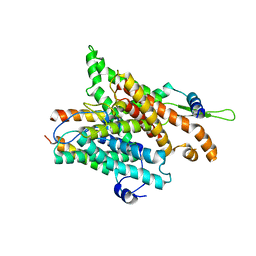 | | The structure of a protein conducting channel | | 分子名称: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, SecBeta | | 著者 | van den Berg, B, Clemons Jr, W.M, Collinson, I, Modis, Y, Hartmann, E, Harrison, S.C, Rapoport, T.A. | | 登録日 | 2003-11-13 | | 公開日 | 2004-01-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | X-ray structure of a protein-conducting channel
Nature, 427, 2004
|
|
1SUV
 
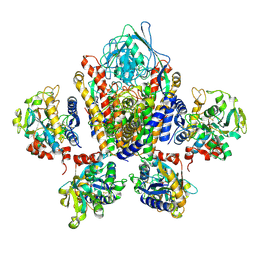 | | Structure of Human Transferrin Receptor-Transferrin Complex | | 分子名称: | CARBONATE ION, FE (III) ION, Serotransferrin, ... | | 著者 | Cheng, Y, Zak, O, Aisen, P, Harrison, S.C, Walz, T. | | 登録日 | 2004-03-26 | | 公開日 | 2004-04-13 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (7.5 Å) | | 主引用文献 | Structure of the Human Transferrin Receptor-Transferrin Complex
Cell(Cambridge,Mass.), 116, 2004
|
|
1T2K
 
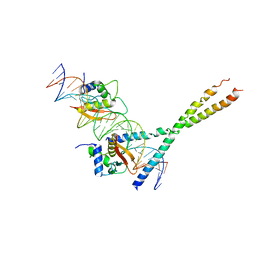 | | Structure Of The DNA Binding Domains Of IRF3, ATF-2 and Jun Bound To DNA | | 分子名称: | 31-MER, Cyclic-AMP-dependent transcription factor ATF-2, Interferon regulatory factor 3, ... | | 著者 | Panne, D, Maniatis, T, Harrison, S.C. | | 登録日 | 2004-04-21 | | 公開日 | 2004-11-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer.
Embo J., 23, 2004
|
|
2B6O
 
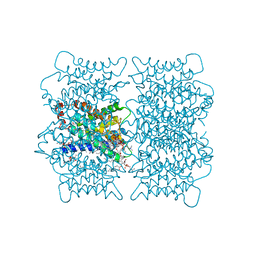 | | Electron crystallographic structure of lens Aquaporin-0 (AQP0) (lens MIP) at 1.9A resolution, in a closed pore state | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Lens fiber major intrinsic protein | | 著者 | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | 登録日 | 2005-10-03 | | 公開日 | 2005-12-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | 主引用文献 | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
3CRO
 
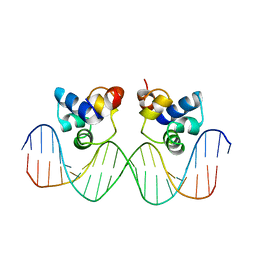 | |
1PER
 
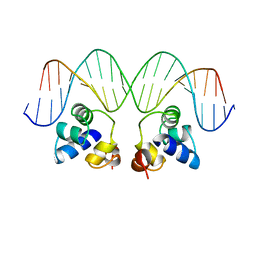 | |
1SVB
 
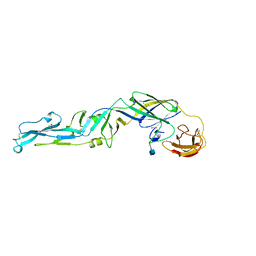 | |
1RHZ
 
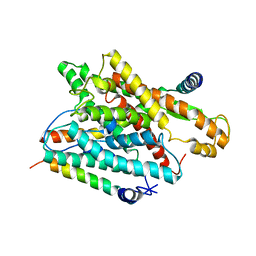 | | The structure of a protein conducting channel | | 分子名称: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, SecBeta | | 著者 | van den Berg, B, Clemons Jr, W.M, Collinson, I, Modis, Y, Hartmann, E, Harrison, S.C, Rapoport, T.A. | | 登録日 | 2003-11-15 | | 公開日 | 2004-01-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | X-ray structure of a protein-conducting channel.
Nature, 427, 2004
|
|
4HKB
 
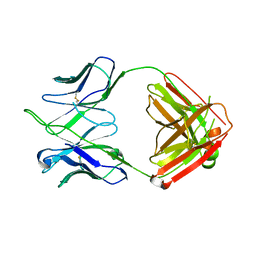 | |
4HK3
 
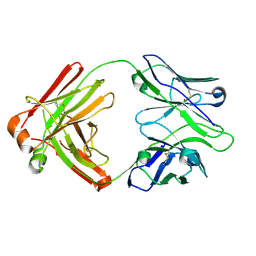 | | I2 Fab (unbound) from CH65-CH67 Lineage | | 分子名称: | I2 heavy chain, I2 light chain | | 著者 | Schmidt, A.G, Harrison, S.C. | | 登録日 | 2012-10-14 | | 公開日 | 2012-11-21 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1RTD
 
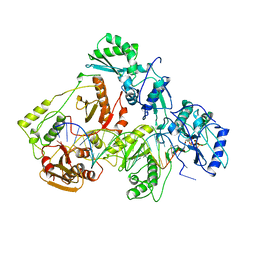 | | STRUCTURE OF A CATALYTIC COMPLEX OF HIV-1 REVERSE TRANSCRIPTASE: IMPLICATIONS FOR NUCLEOSIDE ANALOG DRUG RESISTANCE | | 分子名称: | DNA PRIMER FOR REVERSE TRANSCRIPTASE, DNA TEMPLATE FOR REVERSE TRANSCRIPTASE, MAGNESIUM ION, ... | | 著者 | Chopra, R, Huang, H, Verdine, G.L, Harrison, S.C. | | 登録日 | 1998-08-26 | | 公開日 | 1998-12-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance.
Science, 282, 1998
|
|
2CRO
 
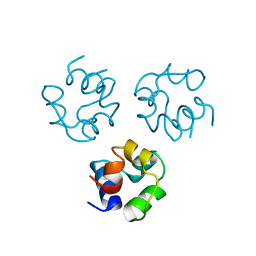 | |
6OJ3
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ4
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
5UK2
 
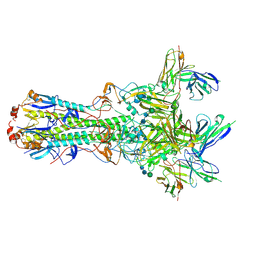 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | 著者 | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | 登録日 | 2017-01-19 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
5UJZ
 
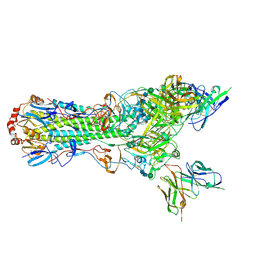 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | 著者 | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | 登録日 | 2017-01-19 | | 公開日 | 2017-05-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
2QUQ
 
 | |
6PP7
 
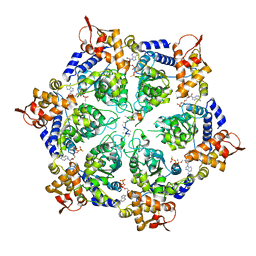 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-05 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
2R7T
 
 | | Crystal Structure of Rotavirus SA11 VP1/RNA (UGUGAACC) Complex | | 分子名称: | RNA (5'-R(*UP*GP*UP*GP*AP*AP*CP*C)-3'), RNA-dependent RNA polymerase | | 著者 | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | 登録日 | 2007-09-10 | | 公開日 | 2008-07-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
2R7O
 
 | | Crystal Structure of VP1 apoenzyme of Rotavirus SA11 (N-terminal hexahistidine-tagged) | | 分子名称: | RNA-dependent RNA polymerase | | 著者 | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | 登録日 | 2007-09-09 | | 公開日 | 2008-07-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
2R7S
 
 | | Crystal Structure of Rotavirus SA11 VP1 / RNA (UGUGCC) complex | | 分子名称: | PHOSPHATE ION, RNA (5'-R(*UP*GP*UP*GP*CP*C)-3'), RNA-dependent RNA polymerase | | 著者 | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | 登録日 | 2007-09-10 | | 公開日 | 2008-07-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.24 Å) | | 主引用文献 | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
6PP5
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 4 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-05 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.98 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP6
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | 登録日 | 2019-07-05 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.28 Å) | | 主引用文献 | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
