7TY8
 
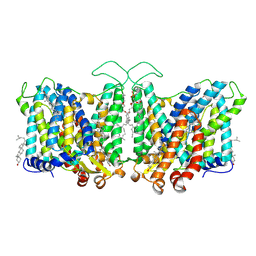 | | Cryo-EM structure of human Anion Exchanger 1 bound to Niflumic Acid | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, ... | | Authors: | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1J2F
 
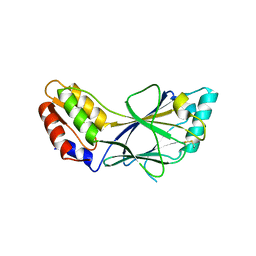 | | X-ray crystal structure of IRF-3 and its functional implications | | Descriptor: | Interferon regulatory factor 3 | | Authors: | Takahasi, K, Noda, N, Horiuchi, M, Mori, M, Okabe, Y, Fukuhara, Y, Terasawa, H, Fujita, T, Inagaki, F. | | Deposit date: | 2003-01-04 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of IRF-3 and its functional implications.
Nat.Struct.Biol., 10, 2003
|
|
1V84
 
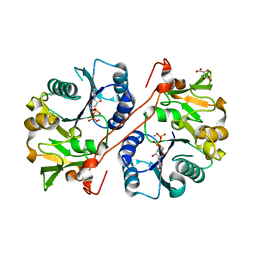 | | Crystal structure of human GlcAT-P in complex with N-acetyllactosamine, Udp, and Mn2+ | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID, MANGANESE (II) ION, ... | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1V83
 
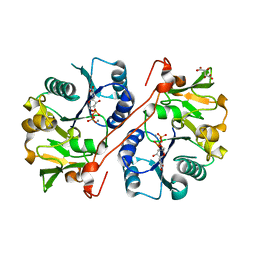 | | Crystal structure of human GlcAT-P in complex with Udp and Mn2+ | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID, MANGANESE (II) ION, ... | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1V82
 
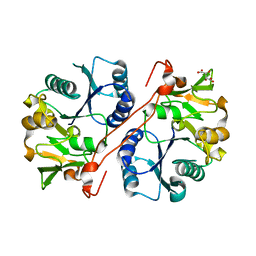 | | Crystal structure of human GlcAT-P apo form | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1HD0
 
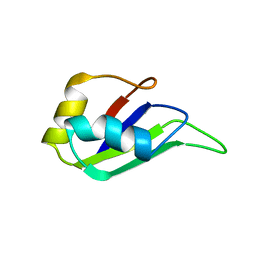 | | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0 (HNRNP D0 RBD1), NMR | | Descriptor: | PROTEIN (HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0) | | Authors: | Nagata, T, Kurihara, Y, Matsuda, G, Saeki, J, Kohno, T, Yanagida, Y, Ishikawa, F, Uesugi, S, Katahira, M. | | Deposit date: | 1999-05-18 | | Release date: | 2000-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0.
J.Mol.Biol., 287, 1999
|
|
1HD1
 
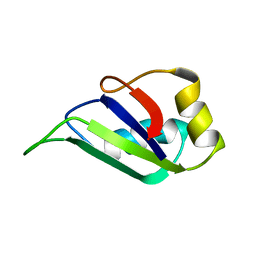 | | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0 (HNRNP D0 RBD1), NMR | | Descriptor: | PROTEIN (HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN D0) | | Authors: | Nagata, T, Kurihara, Y, Matsuda, G, Saeki, J, Kohno, T, Yanagida, Y, Ishikawa, F, Uesugi, S, Katahira, M. | | Deposit date: | 1999-05-18 | | Release date: | 2000-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0.
J.Mol.Biol., 287, 1999
|
|
1IW8
 
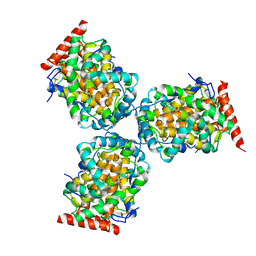 | | Crystal Structure of a mutant of acid phosphatase from Escherichia blattae (G74D/I153T) | | Descriptor: | SULFATE ION, acid phosphatase | | Authors: | Ishikawa, K, Mihara, Y, Shimba, N, Ohtsu, N, Kawasaki, H, Suzuki, E, Asano, Y. | | Deposit date: | 2002-04-22 | | Release date: | 2002-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enhancement of nucleoside phosphorylation activity in an acid phosphatase
PROTEIN ENG., 15, 2002
|
|
1EOI
 
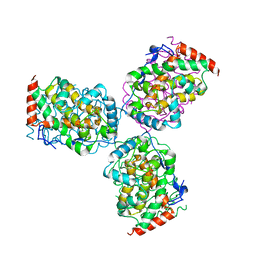 | | CRYSTAL STRUCTURE OF ACID PHOSPHATASE FROM ESCHERICHIA BLATTAE COMPLEXED WITH THE TRANSITION STATE ANALOG MOLYBDATE | | Descriptor: | ACID PHOSPHATASE, MOLYBDATE ION | | Authors: | Ishikawa, K, Mihara, Y, Gondoh, K, Suzuki, E, Asano, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2001-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of a novel acid phosphatase from Escherichia blattae and its complex with the transition-state analog molybdate.
EMBO J., 19, 2000
|
|
5XN8
 
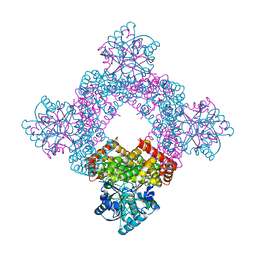 | | Structure of glycerol dehydrogenase crystallised as a contaminant | | Descriptor: | GLYCEROL, Glycerol Dehydrogenase, ZINC ION | | Authors: | Hatti, K, Mathiharan, Y.K, Srinivasan, N, Murthy, M.R.N. | | Deposit date: | 2017-05-19 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Seeing but not believing: the structure of glycerol dehydrogenase initially assumed to be the structure of a survival protein from Salmonella typhimurium
Acta Crystallogr.,Sect.D, 73, 2017
|
|
2LCF
 
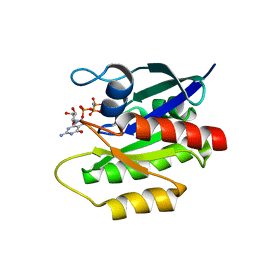 | | Solution structure of GppNHp-bound H-RasT35S mutant protein | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Araki, M, Shima, F, Yoshikawa, Y, Muraoka, S, Ijiri, Y, Nagahara, Y, Shirono, T, Kataoka, T, Tamura, A. | | Deposit date: | 2011-04-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the state 1 conformer of GTP-bound H-Ras protein and distinct dynamic properties between the state 1 and state 2 conformers.
J.Biol.Chem., 286, 2011
|
|
1D2T
 
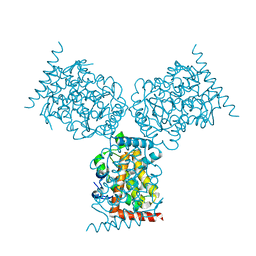 | | CRYSTAL STRUCTURE OF ACID PHOSPHATASE FROM ESCHERICHIA BLATTAE | | Descriptor: | ACID PHOSPHATASE, SULFATE ION | | Authors: | Ishikawa, K, Mihara, Y, Gondoh, K, Suzuki, E, Asano, Y. | | Deposit date: | 1999-09-28 | | Release date: | 2000-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of a novel acid phosphatase from Escherichia blattae and its complex with the transition-state analog molybdate.
EMBO J., 19, 2000
|
|
5YIN
 
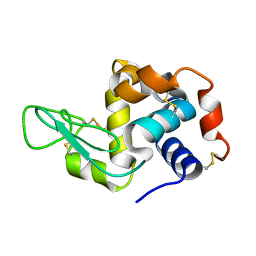 | |
2DQ6
 
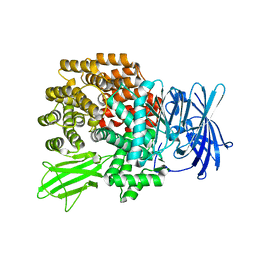 | | Crystal Structure of Aminopeptidase N from Escherichia coli | | Descriptor: | Aminopeptidase N, SULFATE ION, ZINC ION | | Authors: | Nakajima, Y, Onohara, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
1X0R
 
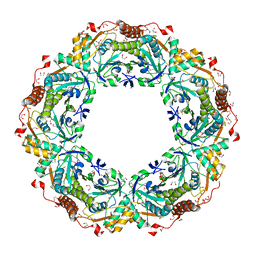 | | Thioredoxin Peroxidase from Aeropyrum pernix K1 | | Descriptor: | 1,2-ETHANEDIOL, Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Inoue, T, Matsumura, H, Kobayashi, A, Hagihara, Y, Uegaki, K, Ataka, M, Kai, Y, Ishikawa, K. | | Deposit date: | 2005-03-28 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of thioredoxin peroxidase from aerobic hyperthermophilic archaeon Aeropyrum pernix K1
Proteins, 62, 2006
|
|
2DCM
 
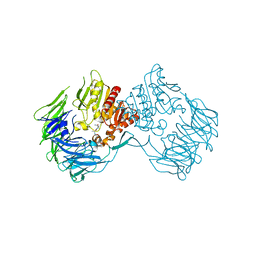 | | The Crystal Structure of S603A Mutated Prolyl Tripeptidyl Aminopeptidase Complexed with Substrate | | Descriptor: | GLYCYLALANYL-N-2-NAPHTHYL-L-PROLINEAMIDE, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2006-01-09 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2D5L
 
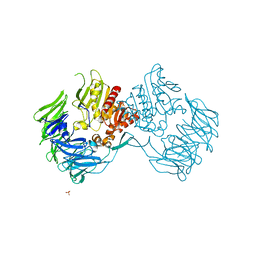 | | Crystal Structure of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis | | Descriptor: | SULFATE ION, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2005-11-02 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
