2VR8
 
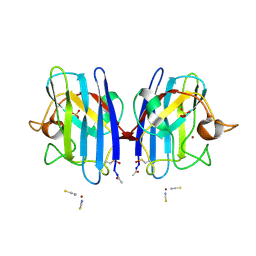 | | Crystal Structure of G85R ALS mutant of Human Cu,Zn Superoxide Dismutase (CuZnSOD) at 1.36 A resolution | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S, Cao, X, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Selverstone Valentine, J, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of the G85R Variant of Sod1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
2VR6
 
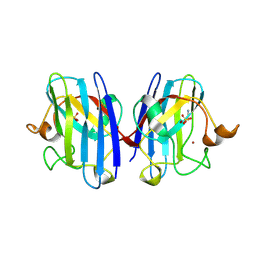 | | Crystal Structure of G85R ALS mutant of Human Cu,Zn Superoxide Dismutase (CuZnSOD) at 1.3 A resolution | | Descriptor: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S, Cao, X, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Selverstone Valentine, J, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the G85R Variant of Sod1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
2AHB
 
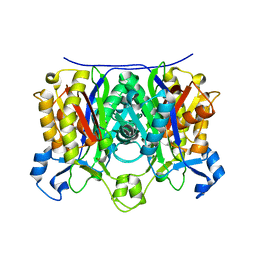 | | X-ray crystal structure of R46A,R161A mutant of Mycobacterium tuberculosis FabH | | Descriptor: | Beta- ketoacyl-ACP synthase III | | Authors: | Brown, A.K, Sridharan, S, Kremer, L, Lindenberg, S, Dover, L.G, Sacchettini, J.C, Besra, G.S. | | Deposit date: | 2005-07-27 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Mechanism of the Mycobacterium tuberculosis {beta}-Ketoacyl-Acyl Carrier Protein Synthase III mtFabH: FACTORS INFLUENCING CATALYSIS AND SUBSTRATE SPECIFICITY
J.Biol.Chem., 280, 2005
|
|
2AJ9
 
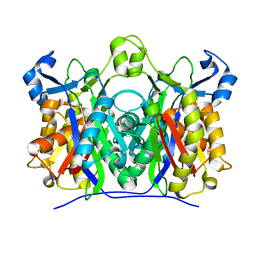 | | X-ray crystal structure of W42A,R161A double mutant of Mycobacterium tuberculosis beta-ketoacyl-ACP synthase III | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase III | | Authors: | Brown, A.K, Sridharan, S, Kremer, L, Lindenberg, S, Dover, L.G, Sacchettini, J.C, Besra, G.S. | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Mechanism of the Mycobacterium tuberculosis beta-Ketoacyl-Acyl Carrier Protein Synthase III mtFabH: Factors Influencing Catalysis And Substrate Specificity.
J.Biol.Chem., 280, 2005
|
|
1C7K
 
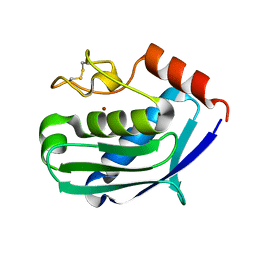 | | CRYSTAL STRUCTURE OF THE ZINC PROTEASE | | Descriptor: | CALCIUM ION, ZINC ENDOPROTEASE, ZINC ION | | Authors: | Kurisu, G, Harada, S, Kai, Y. | | Deposit date: | 2000-02-19 | | Release date: | 2001-04-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the zinc-binding site in the crystal structure of a zinc endoprotease from Streptomyces caespitosus at 1 A resolution.
J.Inorg.Biochem., 82, 2000
|
|
5X2V
 
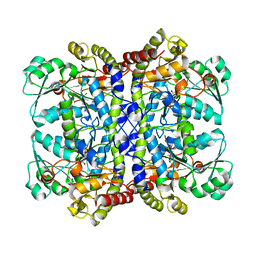 | |
5X2Y
 
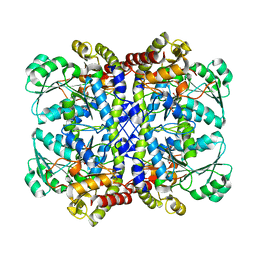 | |
7X0E
 
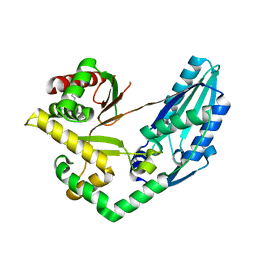 | | Structure of Pseudomonas NRPS protein, AmbB-TC in apo form | | Descriptor: | AMB antimetabolite synthase AmbB, N-methyl-N-[(2S,3R,4R,5R)-2,3,4,5,6-pentakis(oxidanyl)hexyl]nonanamide | | Authors: | ChuYuanKee, M, Bharath, S.R, Song, H. | | Deposit date: | 2022-02-22 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
1ZIA
 
 | | OXIDIZED PSEUDOAZURIN | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystallization and preliminary X-ray studies on pseudoazurin from Achromobacter cycloclastes IAM1013.
J.Biochem.(Tokyo), 114, 1993
|
|
1ZIB
 
 | | REDUCED PSEUDOAZURIN | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Nishio, N, Hamanaka, S, Shimomura, T, Harada, S, Suzuki, S, Kohzuma, T, Shidara, S, Iwasaki, H, Kai, Y. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallization and preliminary X-ray studies on pseudoazurin from Achromobacter cycloclastes IAM1013.
J.Biochem.(Tokyo), 114, 1993
|
|
7ED9
 
 | | Crystal structure of selenomethionine-labeled Thermus thermophilus FakA ATP-binding domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable kinase | | Authors: | Nakatani, M, Nakahara, S, Fukui, K, Murakawa, T, Masui, R. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01764154 Å) | | Cite: | Crystal structure of a nucleotide-binding domain of fatty acid kinase FakA from Thermus thermophilus HB8.
J.Struct.Biol., 214, 2022
|
|
5X2W
 
 | |
5X2Z
 
 | |
7X0F
 
 | |
5X2X
 
 | |
2H5X
 
 | | RuvA from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, Holliday junction ATP-dependent DNA helicase ruvA | | Authors: | Prabu, J.R, Thamotharan, S, Khanduja, J.S, Alipio, E.Z, Kim, C.Y, Waldo, G.S, Terwilliger, T.C, Segelke, B, Lekin, T, Toppani, D, Hung, L.W, Yu, M, Bursey, E, Muniyappa, K, Chandra, N.R, Vijayan, M. | | Deposit date: | 2006-05-28 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Mycobacterium tuberculosis RuvA, a protein involved in recombination.
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
7WMC
 
 | | Crystal structure of macrocyclic peptide 1 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, Peptide1 | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
7WMT
 
 | | Crystal structure of small molecule 13 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, [(2~{R},4~{S})-4-[2-(aminomethyl)imidazol-1-yl]-2-[1-[(4-chlorophenyl)methyl]-5-methyl-indol-2-yl]pyrrolidin-1-yl]-(1~{H}-pyrrolo[2,3-b]pyridin-5-yl)methanone | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
7X17
 
 | | Structure of Pseudomonas NRPS protein, AmbB-TC bound to Ppant-L-Ala | | Descriptor: | AMB antimetabolite synthase AmbB, S-[2-[3-[[(2S)-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butanoyl]amino]propanoylamino]ethyl] (2R)-2-azanylpropanethioate | | Authors: | ChuYuanKee, M, Bharath, S.R, Song, H. | | Deposit date: | 2022-02-23 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
5YJX
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with myxothiazol. | | Descriptor: | (2Z,6E)-7-{2'-[(2E,4E)-1,6-DIMETHYLHEPTA-2,4-DIENYL]-2,4'-BI-1,3-THIAZOL-4-YL}-3,5-DIMETHOXY-4-METHYLHEPTA-2,6-DIENAMID E, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5YJY
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with AC0-12. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-dodecyl-1-oxidanidyl-quinolin-1-ium-4-ol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5ZDP
 
 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with ferulenol | | Descriptor: | 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, Alternative oxidase, mitochondrial, ... | | Authors: | Shiba, T, Inaoka, D.K, Takahashi, G, Tsuge, C, Kido, Y, Young, L, Ueda, S, Balogun, E.O, Nara, T, Honma, T, Tanaka, A, Inoue, M, Saimoto, H, Harada, S, Moore, A.L, Kita, K. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Insights into the ubiquinol/dioxygen binding and proton relay pathways of the alternative oxidase.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
2D22
 
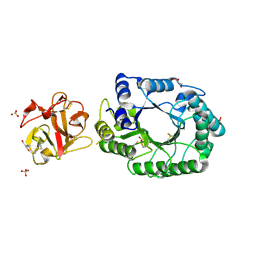 | | Crystal structure of covalent glycosyl-enzyme intermediate of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
5X30
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase C116H mutant with L-homocysteine intermediates. | | Descriptor: | (2E)-2-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}but-2-enoic acid, (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-sulfanyl-butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, ... | | Authors: | Shiba, T, Sato, D, Harada, S. | | Deposit date: | 2017-02-02 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into homocysteine degradation by a mutant of methionine gamma-lyase based on substrate-assisted catalysis
Protein Sci., 26, 2017
|
|
2D23
 
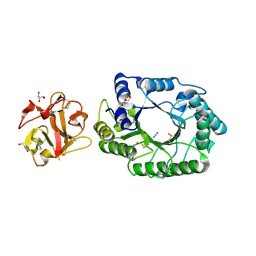 | | Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | AZIDE ION, ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
