7X0E
 
 | | Structure of Pseudomonas NRPS protein, AmbB-TC in apo form | | 分子名称: | AMB antimetabolite synthase AmbB, N-methyl-N-[(2S,3R,4R,5R)-2,3,4,5,6-pentakis(oxidanyl)hexyl]nonanamide | | 著者 | ChuYuanKee, M, Bharath, S.R, Song, H. | | 登録日 | 2022-02-22 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
8I3Y
 
 | | Crystal structure of ASCT from Trypanosoma brucei in complex with Succinyl-CoA. | | 分子名称: | CALCIUM ION, SUCCINIC ACID, SUCCINYL-COENZYME A, ... | | 著者 | Mochizuki, K, Inaoka, D.K, Fukuda, K, Kurasawa, H, Iyoda, K, Nakai, U, Harada, S, Balogun, E.O, Mazet, M, Millerioux, Y, Bringaud, F, Boshart, M, Hirayama, K, Kita, K, Shiba, T. | | 登録日 | 2023-01-18 | | 公開日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of ASCT from Trypanosoma brucei in complex with Succinyl-CoA.
To Be Published
|
|
8I40
 
 | | Crystal structure of ASCT from Trypanosoma brucei in complex with CoA. | | 分子名称: | ACETATE ION, CALCIUM ION, COENZYME A, ... | | 著者 | Mochizuki, K, Inaoka, D.K, Fukuda, K, Kurasawa, H, Iyoda, K, Nakai, U, Harada, S, Balogun, E.O, Mazet, M, Millerioux, Y, Bringaud, F, Boshart, M, Hirayama, K, Kita, K, Shiba, T. | | 登録日 | 2023-01-18 | | 公開日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Crystal structure of ligand complexes of ASCT from Trypanosoma brucei and molecular mechanism in comparison with mammalian SCOT.
To Be Published
|
|
5XX1
 
 | | Crystal structure of Arginine decarboxylase (AdiA) from Salmonella typhimurium | | 分子名称: | Arginine decarboxylase, PHOSPHATE ION | | 著者 | Deka, G, Bharath, S.R, Shavithri, H.S, Murthy, M.R.N. | | 登録日 | 2017-06-30 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural studies on the decameric S. typhimurium arginine decarboxylase (ADC): Pyridoxal 5'-phosphate binding induces conformational changes
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5X30
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase C116H mutant with L-homocysteine intermediates. | | 分子名称: | (2E)-2-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}but-2-enoic acid, (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-sulfanyl-butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, ... | | 著者 | Shiba, T, Sato, D, Harada, S. | | 登録日 | 2017-02-02 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and mechanistic insights into homocysteine degradation by a mutant of methionine gamma-lyase based on substrate-assisted catalysis
Protein Sci., 26, 2017
|
|
4XGM
 
 | |
1X26
 
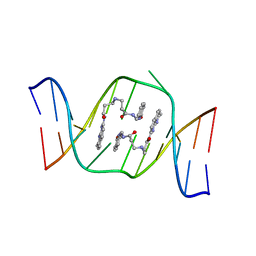 | | Solution structure of the AA-mismatch DNA complexed with naphthyridine-azaquinolone | | 分子名称: | 5'-D(*CP*AP*TP*TP*CP*AP*GP*TP*TP*AP*G)-3', 5'-D(*CP*TP*AP*AP*CP*AP*GP*AP*AP*TP*G)-3', N~3~-{3-[(7-METHYL-1,8-NAPHTHYRIDIN-2-YL)AMINO]-3-OXOPROPYL}-N~1~-[(7-OXO-7,8-DIHYDRO-1,8-NAPHTHYRIDIN-2-YL)METHYL]-BET A-ALANINAMIDE | | 著者 | Nakatani, K, Hagihara, S, Goto, Y, Kobori, A, Hagihara, M, Hayashi, G, Kyo, M, Nomura, M, Mishima, M, Kojima, C. | | 登録日 | 2005-04-20 | | 公開日 | 2006-04-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats
Nat.Chem.Biol., 1, 2005
|
|
4XGL
 
 | |
5YJX
 
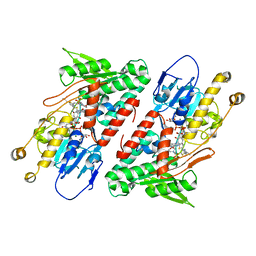 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with myxothiazol. | | 分子名称: | (2Z,6E)-7-{2'-[(2E,4E)-1,6-DIMETHYLHEPTA-2,4-DIENYL]-2,4'-BI-1,3-THIAZOL-4-YL}-3,5-DIMETHOXY-4-METHYLHEPTA-2,6-DIENAMID E, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | 登録日 | 2017-10-11 | | 公開日 | 2018-02-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5YJY
 
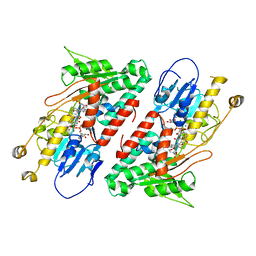 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with AC0-12. | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-dodecyl-1-oxidanidyl-quinolin-1-ium-4-ol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | 登録日 | 2017-10-11 | | 公開日 | 2018-02-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5YJW
 
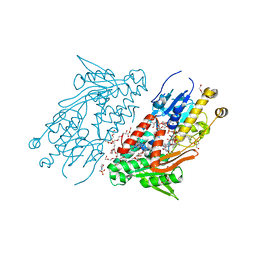 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with the competitive inhibitor, stigmatellin. | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Yamasita, T, Inaoka, D.K, Shiba, T, Oohashi, T, Iwata, S, Yagi, T, Kosaka, H, Harada, S, Kita, K, Hirano, K. | | 登録日 | 2017-10-11 | | 公開日 | 2018-02-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Ubiquinone binding site of yeast NADH dehydrogenase revealed by structures binding novel competitive- and mixed-type inhibitors
Sci Rep, 8, 2018
|
|
5ZDR
 
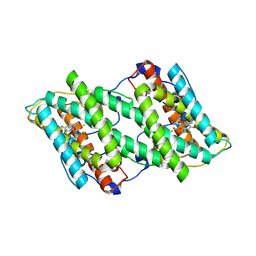 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with ascofuranone derivative | | 分子名称: | 3-chloro-4,6-dihydroxy-5-[(2E,6E,8S)-8-hydroxy-3,7-dimethylnona-2,6-dien-1-yl]-2-methylbenzaldehyde, Alternative oxidase, mitochondrial, ... | | 著者 | Shiba, T, Inaoka, D.K, Takahashi, G, Tsuge, C, Kido, Y, Young, L, Ueda, S, Balogun, E.O, Nara, T, Honma, T, Tanaka, A, Inoue, M, Saimoto, H, Harada, S, Moore, A.L, Kita, K. | | 登録日 | 2018-02-23 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Insights into the ubiquinol/dioxygen binding and proton relay pathways of the alternative oxidase.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
5ZDQ
 
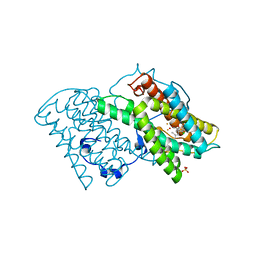 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with COLLETOCHLORIN B | | 分子名称: | 3-chloro-5-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-4,6-dihydroxy-2-methylbenzaldehyde, Alternative oxidase, mitochondrial, ... | | 著者 | Shiba, T, Inaoka, D.K, Takahashi, G, Tsuge, C, Kido, Y, Young, L, Ueda, S, Balogun, E.O, Nara, T, Honma, T, Tanaka, A, Inoue, M, Saimoto, H, Harada, S, Moore, A.L, Kita, K. | | 登録日 | 2018-02-23 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Insights into the ubiquinol/dioxygen binding and proton relay pathways of the alternative oxidase.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
5ZDP
 
 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with ferulenol | | 分子名称: | 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, Alternative oxidase, mitochondrial, ... | | 著者 | Shiba, T, Inaoka, D.K, Takahashi, G, Tsuge, C, Kido, Y, Young, L, Ueda, S, Balogun, E.O, Nara, T, Honma, T, Tanaka, A, Inoue, M, Saimoto, H, Harada, S, Moore, A.L, Kita, K. | | 登録日 | 2018-02-23 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Insights into the ubiquinol/dioxygen binding and proton relay pathways of the alternative oxidase.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
5X2W
 
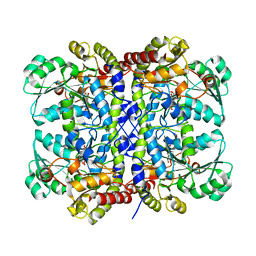 | |
5X2Z
 
 | |
5X2X
 
 | |
7ED9
 
 | | Crystal structure of selenomethionine-labeled Thermus thermophilus FakA ATP-binding domain | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable kinase | | 著者 | Nakatani, M, Nakahara, S, Fukui, K, Murakawa, T, Masui, R. | | 登録日 | 2021-03-15 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.01764154 Å) | | 主引用文献 | Crystal structure of a nucleotide-binding domain of fatty acid kinase FakA from Thermus thermophilus HB8.
J.Struct.Biol., 214, 2022
|
|
7UN5
 
 | |
7UMQ
 
 | |
6AJ6
 
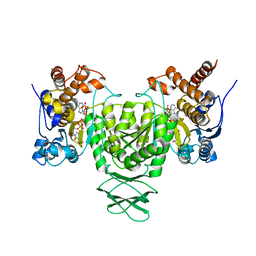 | | Crystal structure of Trypanosoma brucei glycosomal isocitrate dehydrogenase in complex with NADP+ | | 分子名称: | Isocitrate dehydrogenase [NADP], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | 登録日 | 2018-08-27 | | 公開日 | 2019-08-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
5X2V
 
 | |
5X2Y
 
 | |
7ED6
 
 | | Crystal structure of Thermus thermophilus FakA ATP-binding domain | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable kinase | | 著者 | Nakatani, M, Nakahara, S, Fukui, K, Murakawa, T, Masui, R. | | 登録日 | 2021-03-15 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-01-11 | | 実験手法 | X-RAY DIFFRACTION (1.92850327 Å) | | 主引用文献 | Crystal structure of a nucleotide-binding domain of fatty acid kinase FakA from Thermus thermophilus HB8.
J.Struct.Biol., 214, 2022
|
|
7CII
 
 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with L- methionine methyl ester (external aldimine form). | | 分子名称: | L-methionine decarboxylase, methyl (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-4-methylsulfanyl-butanoate | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
