2PCQ
 
 | | Crystal structure of putative dihydrodipicolinate synthase (TTHA0737) from Thermus Thermophilus HB8 | | Descriptor: | GLYCEROL, POTASSIUM ION, Putative dihydrodipicolinate synthase | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Kitamura, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of putative dihydrodipicolinate synthase (TTHA0737) from Thermus Thermophilus HB8
To be Published
|
|
5AVM
 
 | | Crystal structures of 5-aminoimidazole ribonucleotide (AIR) synthetase, PurM, from Thermus thermophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Watanabe, Y, Nakagawa, N, Ebihara, A, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 159, 2016
|
|
2P2O
 
 | | Crystal structure of maltose transacetylase from Geobacillus kaustophilus P2(1) crystal form | | Descriptor: | Maltose transacetylase | | Authors: | Liu, Z.J, Li, Y, Chen, L, Zhu, J, Rose, J.P, Ebihara, A, Yokoyama, S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of Maltose Transacetylase from Geobacillus Kaustophilus at 1.8 Angstrom Resolution
To be Published
|
|
2DQB
 
 | | Crystal structure of dNTP triphosphohydrolase from Thermus thermophilus HB8, which is homologous to dGTP triphosphohydrolase | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, putative, MAGNESIUM ION | | Authors: | Kondo, N, Nakagawa, N, Ebihara, A, Chen, L, Liu, Z.-J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-25 | | Release date: | 2007-01-23 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of dNTP-inducible dNTP triphosphohydrolase: insight into broad specificity for dNTPs and triphosphohydrolase-type hydrolysis
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2ECR
 
 | | Crystal structure of the ligand-free form of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-13 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2DUY
 
 | | Crystal structure of competence protein ComEA-related protein from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, Competence protein ComEA-related protein | | Authors: | Niwa, H, Shimada, A, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-27 | | Release date: | 2007-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of competence protein ComEA-related protein from Thermus thermophilus HB8
To be Published
|
|
2EGZ
 
 | | Crystal structure of the 3-dehydroquinate dehydratase from Aquifex aeolicus VF5 | | Descriptor: | 3-dehydroquinate dehydratase, L(+)-TARTARIC ACID | | Authors: | Karthe, P, Kumarevel, T.S, Ebihara, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-02 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the 3-dehydroquinate dehydratase from Aquifex aeolicus VF5
To be Published
|
|
2EJ5
 
 | | Crystal structure of GK2038 protein (enoyl-CoA hydratase subunit II) from Geobacillus kaustophilus | | Descriptor: | Enoyl-CoA hydratase subunit II | | Authors: | Okazaki, N, Agari, Y, Ebihara, A, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GK2038 protein (enoyl-CoA hydratase subunit II) from Geobacillus kaustophilus
To be Published
|
|
8A11
 
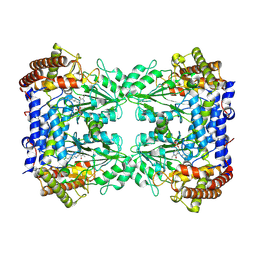 | | Cryo-EM structure of the Human SHMT1-RNA complex | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, cytosolic | | Authors: | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 2024
|
|
8R7H
 
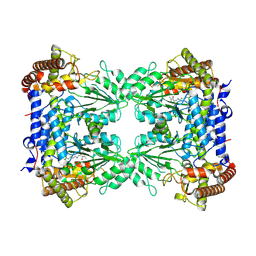 | | Cryo-EM structure of Human SHMT1 | | Descriptor: | Serine hydroxymethyltransferase, cytosolic | | Authors: | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | Deposit date: | 2023-11-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 84, 2024
|
|
8BV0
 
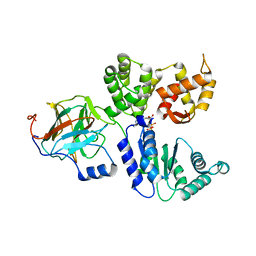 | | Binary complex between the NB-ARC domain from the Tomato immune receptor NRC1 and the SPRY domain-containing effector SS15 from the potato cyst nematode | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NRC1, Truncated secreted SPRY domain-containing protein 15 (Fragment) | | Authors: | Contreras, M.P, Pai, H, Muniyandi, S, Toghani, A, Lawson, D.M, Tumtas, Y, Duggan, C, Yuen, E.L.H, Stevenson, C.E.M, Harant, A, Wu, C.H, Bozkurt, T.O, Kamoun, S, Derevnina, L. | | Deposit date: | 2022-12-01 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Resurrection of plant disease resistance proteins via helper NLR bioengineering.
Sci Adv, 9, 2023
|
|
8WDU
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by sucrose density | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
8WDV
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
6LTB
 
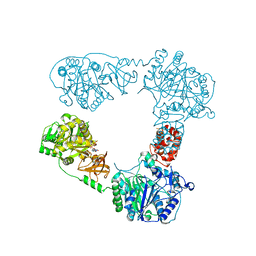 | |
6LTA
 
 | |
6LTD
 
 | |
6LTC
 
 | |
8JQ5
 
 | |
8JQ4
 
 | |
8JQ6
 
 | |
8JQ3
 
 | |
5ZFS
 
 | | Crystal structure of Arthrobacter globiformis M30 sugar epimerase which can produce D-allulose from D-fructose | | Descriptor: | ACETATE ION, D-allulose-3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Gullapalli, P.K, Ohtani, K, Akimitsu, K, Izumori, K, Kamitori, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray structure of Arthrobacter globiformis M30 ketose 3-epimerase for the production of D-allulose from D-fructose.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4Z5T
 
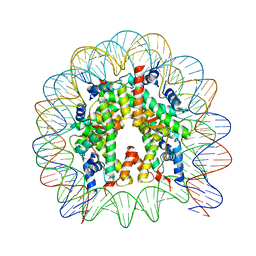 | | The nucleosome containing human H3.5 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Urahama, T, Harada, A, Maehara, K, Horikoshi, N, Sato, K, Sato, Y, Shiraishi, K, Sugino, N, Osakabe, A, Tachiwana, H, Kagawa, W, Kimura, H, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-04-03 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Histone H3.5 forms an unstable nucleosome and accumulates around transcription start sites in human testis.
Epigenetics Chromatin, 9, 2016
|
|
1WXX
 
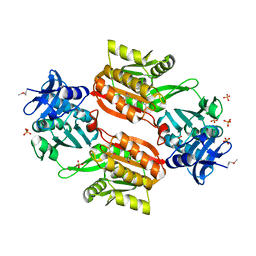 | | Crystal structure of Tt1595, a putative SAM-dependent methyltransferase from Thermus thermophillus HB8 | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, hypothetical protein TTHA1280 | | Authors: | Pioszak, A.A, Murayama, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-02 | | Release date: | 2005-08-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of a putative RNA 5-methyluridine methyltransferase, Thermus thermophilus TTHA1280, and its complex with S-adenosyl-L-homocysteine.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
7WRR
 
 | |
