4D5P
 
 | | Hypocrea jecorina cellobiohydrolase Cel7A E217Q soaked with xylopentaose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, COBALT (II) ION, ... | | Authors: | Momeni, M.H, Ubhayasekera, W, Stahlberg, J, Hansson, H. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Insights Into the Inhibition of Cellobiohydrolase Cel7A by Xylooligosaccharides.
FEBS J., 282, 2015
|
|
4D5J
 
 | | Hypocrea jecorina cellobiohydrolase Cel7A E217Q soaked with xylotriose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, COBALT (II) ION, ... | | Authors: | Momeni, M.H, Ubhayasekera, W, Stahlberg, J, Hansson, H. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights Into the Inhibition of Cellobiohydrolase Cel7A by Xylooligosaccharides.
FEBS J., 282, 2015
|
|
3ZYZ
 
 | | Crystal structure of a glycoside hydrolase family 3 beta-glucosidase, Bgl1 from Hypocrea jecorina at 2.1A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCOSIDE GLUCOHYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandgren, M, Kaper, T, Mikkelsen, N.E, Hansson, H, Piens, K, Gudmundsson, M, Larenas, E, Kelemen, B, Karkehabadi, S. | | Deposit date: | 2011-08-30 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Crystal Structures of a Fungal Family 3 Beta-Glucosidase, Cel3A from Hypocrea Jecorina.
J.Biol.Chem., 289, 2014
|
|
4AX7
 
 | | Hypocrea jecorina Cel6A D221A mutant soaked with 4-Methylumbelliferyl- beta-D-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
3ZYP
 
 | | Cellulose induced protein, Cip1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CIP1, ... | | Authors: | Jacobson, F, Karkehabadi, S, Hansson, H, Goedegebuur, F, Wallace, L, Mitchinson, C, Piens, K, Stals, I, Sandgren, M. | | Deposit date: | 2011-08-24 | | Release date: | 2012-09-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of the Core Domain of a Cellulose Induced Protein (Cip1) from Hypocrea Jecorina, at 1.5 A Resolution.
Plos One, 8, 2013
|
|
3ZZ1
 
 | | Crystal structure of a glycoside hydrolase family 3 beta-glucosidase, Bgl1 from Hypocrea jecorina at 2.1A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCOSIDE GLUCOHYDROLASE, GLYCEROL | | Authors: | Sandgren, M, Kaper, T, Mikkelsen, N.E, Hansson, H, Piens, K, Gudmundsson, M, Larenas, E, Kelemen, B, Karkehabadi, S. | | Deposit date: | 2011-08-31 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Crystal Structures of a Fungal Family 3 Beta-Glucosidase, Cel3A from Hypocrea Jecorina.
J.Biol.Chem., 289, 2014
|
|
4B4H
 
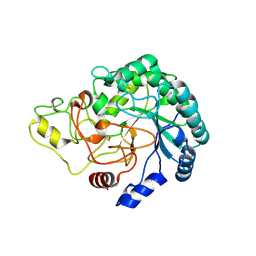 | | Thermobifida fusca cellobiohydrolase Cel6B(E3) catalytic domain | | Descriptor: | BETA-1,4-EXOCELLULASE | | Authors: | Sandgren, M, Wu, M, Stahlberg, J, Karkehabadi, S, Mitchinson, C, Kelemen, B.R, Larenas, E.A, Hansson, H. | | Deposit date: | 2012-07-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of a Bacterial Cellobiohydrolase: The Catalytic Core of the Thermobifida Fusca Family Gh6 Cellobiohydrolase Cel6B.
J.Mol.Biol., 425, 2013
|
|
4B4F
 
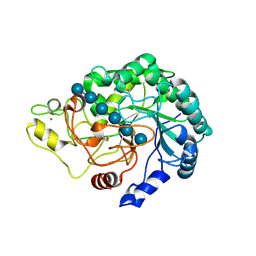 | | Thermobifida fusca Cel6B(E3) co-crystallized with cellobiose | | Descriptor: | BETA-1,4-EXOCELLULASE, CALCIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Sandgren, M, Wu, M, Stahlberg, J, Karkehabadi, S, Mitchinson, C, Kelemen, B.R, Larenas, E.A, Hansson, H. | | Deposit date: | 2012-07-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of a Bacterial Cellobiohydrolase: The Catalytic Core of the Thermobifida Fusca Family Gh6 Cellobiohydrolase Cel6B.
J.Mol.Biol., 425, 2013
|
|
4AVN
 
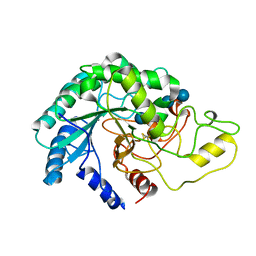 | | Thermobifida fusca cellobiohydrolase Cel6B catalytic mutant D226A- S232A cocrystallized with cellobiose | | Descriptor: | CALCIUM ION, CELLOBIOHYDROLASE. GLYCOSYL HYDROLASE FAMILY 6, beta-D-glucopyranose, ... | | Authors: | Wu, M, Vuong, T.V, Wilson, D.B, Sandgren, M, Stahlberg, J, Hansson, H. | | Deposit date: | 2012-05-28 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Loop Motions Important to Product Expulsion in the Thermobifida Fusca Glycoside Hydrolase Family 6 Cellobiohydrolase from Structural and Computational Studies.
J.Biol.Chem., 288, 2013
|
|
4AVO
 
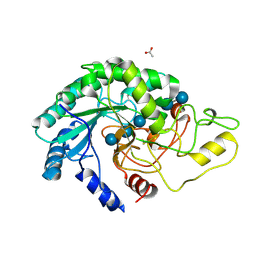 | | Thermobifida fusca cellobiohydrolase Cel6B catalytic mutant D274A cocrystallized with cellobiose | | Descriptor: | ACETATE ION, BETA-1,4-EXOCELLULASE, CALCIUM ION, ... | | Authors: | Wu, M, Vuong, T.V, Wilson, D.B, Sandgren, M, Stahlberg, J, Hansson, H. | | Deposit date: | 2012-05-28 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Loop Motions Important to Product Expulsion in the Thermobifida Fusca Glycoside Hydrolase Family 6 Cellobiohydrolase from Structural and Computational Studies.
J.Biol.Chem., 288, 2013
|
|
6SC9
 
 | | dAb3/HOIP-RBR-HOIPIN-8 | | Descriptor: | 2-[3-[2,6-bis(fluoranyl)-4-(1~{H}-pyrazol-4-yl)phenyl]-3-oxidanylidene-prop-1-enyl]-4-(1-methylpyrazol-4-yl)benzoic acid, CHLORIDE ION, E3 ubiquitin-protein ligase RNF31, ... | | Authors: | Tsai, Y.-C.I, Johansson, H, House, D, Rittinger, K. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Single-Domain Antibodies as Crystallization Chaperones to Enable Structure-Based Inhibitor Development for RBR E3 Ubiquitin Ligases.
Cell Chem Biol, 27, 2020
|
|
6SC8
 
 | | dAb3/HOIP-RBR-Ligand4 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF31, SULFATE ION, ... | | Authors: | Tsai, Y.-C.I, Johansson, H, House, D, Rittinger, K. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Single-Domain Antibodies as Crystallization Chaperones to Enable Structure-Based Inhibitor Development for RBR E3 Ubiquitin Ligases.
Cell Chem Biol, 27, 2020
|
|
6SC7
 
 | | dAb3/HOIP-RBR-Ligand3 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF31, SULFATE ION, ... | | Authors: | Tsai, Y.-C.I, Johansson, H, House, D, Rittinger, K. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Single-Domain Antibodies as Crystallization Chaperones to Enable Structure-Based Inhibitor Development for RBR E3 Ubiquitin Ligases.
Cell Chem Biol, 27, 2020
|
|
6SC5
 
 | | dAb3/HOIP-RBR-Ligand2 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF31, SULFATE ION, ... | | Authors: | Tsai, Y.-C.I, Johansson, H, House, D, Rittinger, K. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-Domain Antibodies as Crystallization Chaperones to Enable Structure-Based Inhibitor Development for RBR E3 Ubiquitin Ligases.
Cell Chem Biol, 27, 2020
|
|
2EW4
 
 | | Solution structure of MrIA | | Descriptor: | MrIA | | Authors: | Nilsson, K.P, Lovelace, E.S, Caesar, C.E, Tynngard, N, Alewood, P.F, Johansson, H.M, Sharpe, I.A, Lewis, R.J, Daly, N.L, Craik, D.J. | | Deposit date: | 2005-11-02 | | Release date: | 2005-11-15 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Solution structure of chi-conopeptide MrIA, a modulator of the human norepinephrine transporter
Biopolymers, 80, 2005
|
|
