1Z38
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with inosine | | Descriptor: | INOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z35
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
4H6P
 
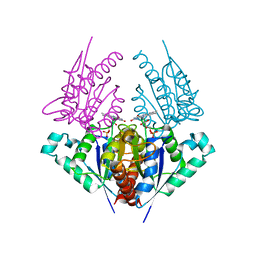 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a R101A substitution. | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Zhang, Y, Robinson, H, Buchko, G.W. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.556 Å) | | Cite: | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
1Z34
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
5B3Z
 
 | |
4KMH
 
 | |
4KM9
 
 | |
1Z37
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with adenosine | | Descriptor: | ADENOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
5B3X
 
 | |
5B3P
 
 | |
4KMA
 
 | |
5BMY
 
 | |
4GKU
 
 | |
1Z36
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
4HS4
 
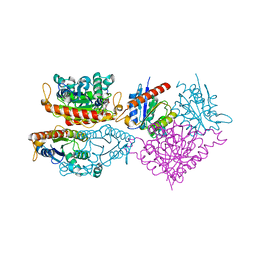 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a Y129N substitution. | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Zhang, Y, Robinson, H, Buchko, G.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
4R7H
 
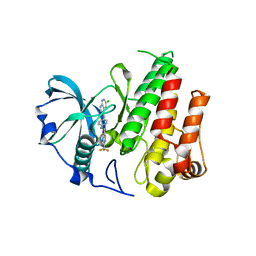 | | Crystal structure of FMS KINASE domain with a small molecular inhibitor, PLX3397 | | Descriptor: | 5-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-{[6-(trifluoromethyl)pyridin-3-yl]methyl}pyridin-2-amine, Macrophage colony-stimulating factor 1 receptor | | Authors: | Zhang, Y, Zhang, K, Zhang, C. | | Deposit date: | 2014-08-27 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8001 Å) | | Cite: | Structure-Guided Blockade of CSF1R Kinase in Tenosynovial Giant-Cell Tumor.
N Engl J Med, 373, 2015
|
|
4R7I
 
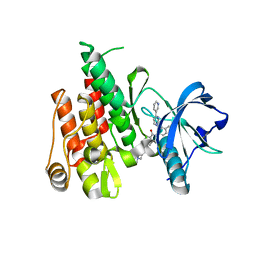 | |
2MNY
 
 | | NMR Structure of KDM5B PHD1 finger | | Descriptor: | Lysine-specific demethylase 5B, ZINC ION | | Authors: | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | Deposit date: | 2014-04-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
3ZEE
 
 | | Electron cyro-microscopy helical reconstruction of Par-3 N terminal domain | | Descriptor: | PARTITIONING DEFECTIVE 3 HOMOLOG | | Authors: | Zhang, Y, Wang, W, Chen, J, Zhang, K, Gao, F, Gong, W, Zhang, M, Sun, F, Feng, W. | | Deposit date: | 2012-12-05 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Intrinsic Self-Assembly of Par-3 N-Terminal Domain.
Structure, 21, 2013
|
|
5XS9
 
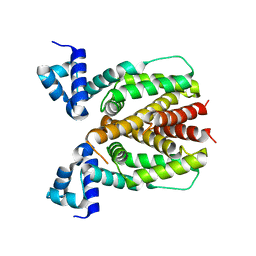 | | Crystal structure of Mycobacterium smegmatis BioQ | | Descriptor: | TetR family transcriptional regulator | | Authors: | Zhang, Y, Ji, Q, Feng, Y. | | Deposit date: | 2017-06-13 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of BioQ suggests a distinct regulatory mechanism for biotin, a nutritional virulence factor in Mycobacterium
To Be Published
|
|
4OIP
 
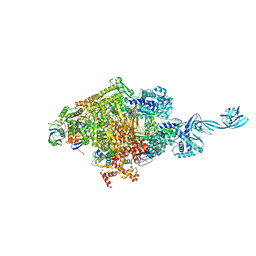 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
5X87
 
 | |
4OIR
 
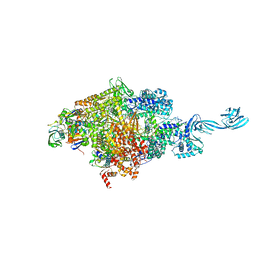 | | Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
6J1L
 
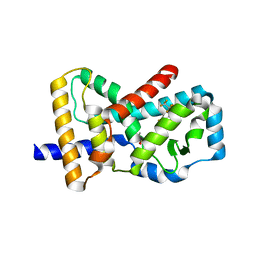 | | Crystal Structure Analysis of the ROR gamma(C455E) | | Descriptor: | 2-[4-(ethylsulfonyl)phenyl]-N-[2'-fluoro-4'-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)[1,1'-biphenyl]-4-yl]acetamide, Nuclear receptor ROR-gamma | | Authors: | zhang, Y, Li, C.C, wu, X.S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Characterization of XY101, a Potent, Selective, and Orally Bioavailable ROR gamma Inverse Agonist for Treatment of Castration-Resistant Prostate Cancer.
J.Med.Chem., 62, 2019
|
|
5GNV
 
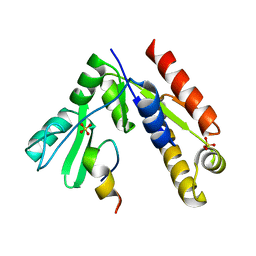 | | Structure of PSD-95/MAP1A complex reveals unique target recognition mode of MAGUK GK domain | | Descriptor: | Disks large homolog 4, Microtubule-associated protein 1A, SULFATE ION | | Authors: | Shang, Y, Xia, Y, Zhu, R, Zhu, J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structure of the PSD-95/MAP1A complex reveals a unique target recognition mode of the MAGUK GK domain
Biochem. J., 474, 2017
|
|
