1FBG
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-mannitol, FRUCTOSE 1,6-BISPHOSPHATASE, MANGANESE (II) ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1RBZ
 
 | | Human GAR Tfase complex structure with polyglutamated 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-4-(2,4-DIAMINO-6-OXO-1,6-DIHYDRO-PYRIMIDIN-5-YL)-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXY-ETHYL)-BUT-2-YL-BENZOYL}-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GLUTAMIC ACID, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
1T8S
 
 | |
4KM8
 
 | | Crystal structure of Sufud60 | | Descriptor: | Suppressor of fused homolog | | Authors: | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | Deposit date: | 2013-05-08 | | Release date: | 2013-11-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
6O3X
 
 | | Crystal structure of yeast Nrd1 CID in complex with Sen1 NIM2 | | Descriptor: | CHLORIDE ION, Helicase SEN1, Protein NRD1 | | Authors: | Zhang, Y, Tong, L. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Identification of Three Sequence Motifs in the Transcription Termination Factor Sen1 that Mediate Direct Interactions with Nrd1.
Structure, 27, 2019
|
|
4Z69
 
 | | Human serum albumin complexed with palmitic acid and diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, PALMITIC ACID, PENTADECANOIC ACID, ... | | Authors: | Zhang, Y, Yang, F. | | Deposit date: | 2015-04-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural basis of non-steroidal anti-inflammatory drug diclofenac binding to human serum albumin.
Chem.Biol.Drug Des., 86, 2015
|
|
6AK5
 
 | | Binary Complex of Human DNA Polymerase Mu with MnGTP | | Descriptor: | DNA-directed DNA/RNA polymerase mu, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
4FK3
 
 | | B-Raf Kinase V600E Oncogenic Mutant in Complex with PLX3203 | | Descriptor: | N-{2,4-difluoro-3-[(5-pyridin-3-yl-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]phenyl}ethanesulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Wang, W, Zhang, K.Y.J. | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3LZC
 
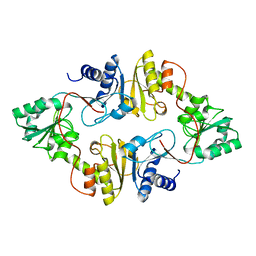 | | Crystal structure of Dph2 from Pyrococcus horikoshii | | Descriptor: | Dph2 | | Authors: | Zhang, Y, Zhu, X, Torelli, A.T, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
5LG4
 
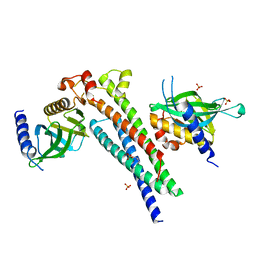 | |
5E17
 
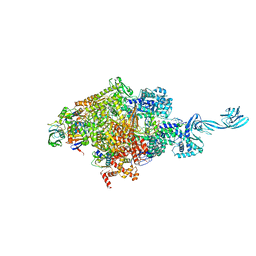 | |
8CZJ
 
 | | A bacteria Zrt/Irt-like protein in the apo state | | Descriptor: | Putative membrane protein, SULFATE ION, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Zhang, Y, Hu, J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structural insights into the elevator-type transport mechanism of a bacterial ZIP metal transporter.
Nat Commun, 14, 2023
|
|
1RBY
 
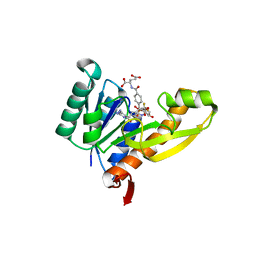 | | Human GAR Tfase complex structure with 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid and substrate beta-GAR | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
6AEC
 
 | | Binary complex of human DNA Polymerase Mu with MnATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed DNA/RNA polymerase mu, MANGANESE (II) ION, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6AK9
 
 | | Pre-catalytic Ternary Complex of Human DNA Polymerase Mu with Templating Cytosine and Incoming Ca-8oxodGTP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6AK8
 
 | | Pre-catalytic Ternary Complex of Human DNA Polymerase Mu with Templating Adenine and Incoming Ca-8oxodGTP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
2LD5
 
 | |
2LLO
 
 | |
4EM0
 
 | |
1T8R
 
 | |
4OXW
 
 | |
1T8Y
 
 | |
4KMH
 
 | |
4KM9
 
 | |
5WMG
 
 | | N-terminal bromodomain of BRD4 in complex with OTX-015 | | Descriptor: | 1,2-ETHANEDIOL, 4-{6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1S)-1-(pyridin-2-yl)ethyl]-1H-pyrrolo[3,2-b]pyridin-3-yl}benzoic acid, Bromodomain-containing protein 4 | | Authors: | Zhang, Y. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
