7W6C
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W69
 
 | | Crystal structure of a PSH1 mutant in complex with EDO | | Descriptor: | 1,2-ETHANEDIOL, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6O
 
 | | Crystal structure of a PSH1 in complex with J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6Q
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
6NT8
 
 | | Cryo-EM structure of full-length chicken STING in the cGAMP-bound tetrameric state | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6NT6
 
 | | Cryo-EM structure of full-length chicken STING in the apo state | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | Deposit date: | 2019-01-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
7XL5
 
 | | Crystal structure of the H42T/A85G/I86A mutant of a nadp-dependent alcohol dehydrogenase | | Descriptor: | NADP-dependent isopropanol dehydrogenase | | Authors: | Jiang, Y.Y, Qu, G, Li, X, Sun, Z.T, Han, X, Liu, W.D. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Engineering the hydrogen transfer pathway of an alcohol dehydrogenase to increase activity by rational enzyme design
Mol Catal, 530, 2022
|
|
6CDY
 
 | | Crystal structure of TEAD complexed with its inhibitor | | Descriptor: | 2-[(4H-1,2,4-triazol-3-yl)sulfanyl]-N-{4-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]phenyl}acetamide, Transcriptional enhancer factor TEF-4 | | Authors: | LIU, S, HAN, X, LUO, X. | | Deposit date: | 2018-02-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Lats1/2 Sustain Intestinal Stem Cells and Wnt Activation through TEAD-Dependent and Independent Transcription.
Cell Stem Cell, 26, 2020
|
|
1WPQ
 
 | | Ternary Complex Of Glycerol 3-phosphate Dehydrogenase 1 with NAD and dihydroxyactone | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic, ... | | Authors: | Ou, X, Han, X, Rao, Z. | | Deposit date: | 2004-09-10 | | Release date: | 2006-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Human Glycerol 3-phosphate Dehydrogenase 1 (GPD1)
J.Mol.Biol., 357, 2006
|
|
8WDM
 
 | | Crystal structure of a novel PU plastic degradation enzyme from Thermaerobacter marianensis | | Descriptor: | Carboxylic ester hydrolase | | Authors: | Li, Z.S, Wang, H, Gao, J, Chen, Y.Y, Wei, H.L, Li, Q, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2023-09-15 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a novel PU plastic degradation enzyme from Thermaerobacter marianensis
To Be Published
|
|
8WDW
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 from uncultured bacterium | | Descriptor: | GLYCEROL, SULFATE ION, UMG-SP2, ... | | Authors: | Cong, L, Li, Z.S, Gao, J, Li, Q, Chen, Y.Y, Han, X, Gert, W, Wei, R, Liu, W.D, Bornscheuer, U.T. | | Deposit date: | 2023-09-16 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-Guided Engineering of a Versatile Urethanase Improves Its Polyurethane Depolymerization Activity.
Adv Sci, 12, 2025
|
|
7SII
 
 | | Human STING bound to both cGAMP and 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide (Compound 53) | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, Stimulator of interferon genes protein, cGAMP | | Authors: | Lu, D, Shang, G, Jie, L, Lu, Y, Bai, X.C, Zhang, X. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Activation of STING by targeting a pocket in the transmembrane domain.
Nature, 604, 2022
|
|
3WUB
 
 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
8XTC
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 mutant from uncultured bacterium in complex with ligand | | Descriptor: | 4-oxidanylbutyl ~{N}-(4-aminophenyl)carbamate, GLYCEROL, umgsp2-mut | | Authors: | Cong, L, Li, Z.S, Zheng, Z.R, Han, X, Gert, W, Wei, R, Liu, W.D, Bornscheuer, U.T. | | Deposit date: | 2024-01-10 | | Release date: | 2025-01-15 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Guided Engineering of a Versatile Urethanase Improves Its Polyurethane Depolymerization Activity.
Adv Sci, 12, 2025
|
|
8XTB
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 from uncultured bacterium in complex with ligand | | Descriptor: | 4-oxidanylbutyl ~{N}-(4-aminophenyl)carbamate, umgsp2 | | Authors: | Cong, L, Li, Z.S, Zheng, Z.R, Han, X, Wei, R, Liu, W.D, Uwe, B. | | Deposit date: | 2024-01-10 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 mutant from uncultured bacterium
To Be Published
|
|
8XRZ
 
 | | Crystal structure of a novel PU plastic degradation enzyme with ligand from Thermaerobacter marianensis | | Descriptor: | 4-oxidanylbutyl ~{N}-[4-[(4-aminophenyl)methyl]phenyl]carbamate, Carboxylic ester hydrolase, SULFATE ION | | Authors: | Li, Z.S, Wang, H, Zheng, Z.R, Cong, L, Chen, Y.Y, Han, X, Wei, R, Uwe, B, liu, W.D. | | Deposit date: | 2024-01-08 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of a novel PU plastic degradation enzyme with ligand from Thermaerobacter marianensis
To Be Published
|
|
3WUF
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUE
 
 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUG
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
8R64
 
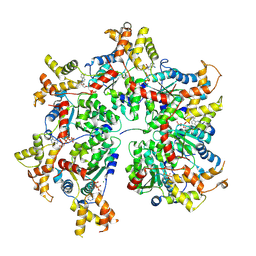 | | Cryo-EM structure of the FIGNL1 AAA hexamer bound to RAD51 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA repair protein RAD51 homolog 1, ... | | Authors: | Carver, A, Yates, L.A, Zhang, X. | | Deposit date: | 2023-11-20 | | Release date: | 2024-09-04 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of FIGNL1 in dissociating RAD51 from DNA and chromatin.
Science, 387, 2025
|
|
8RE4
 
 | |
8RED
 
 | |
8REE
 
 | |
8REB
 
 | |
8REC
 
 | |
