4YY1
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) in complex with human receptor analog 6'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
1KD6
 
 | | Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II | | Descriptor: | EQUINATOXIN II | | Authors: | Hinds, M.G, Zhang, W, Anderluh, G, Hansen, P.E, Norton, R.S. | | Deposit date: | 2001-11-12 | | Release date: | 2002-02-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II: implications for pore formation.
J.Mol.Biol., 315, 2002
|
|
4YY9
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) at 2.6 Angstroms resolution
To Be Published
|
|
4YY7
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) in complex with avian receptor analog 3'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
4YYA
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with avian receptor analog 3'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
4YYB
 
 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with human receptor analog 6'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) at 2.6 Angstroms resolution
To Be Published
|
|
7U0N
 
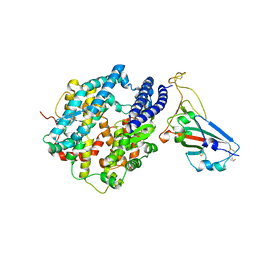 | | Crystal structure of chimeric omicron RBD (strain BA.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Q, Shi, K, Ye, G, Zhang, W, Aihara, H, Li, F. | | Deposit date: | 2022-02-18 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for Human Receptor Recognition by SARS-CoV-2 Omicron Variant BA.1.
J.Virol., 96, 2022
|
|
5VCI
 
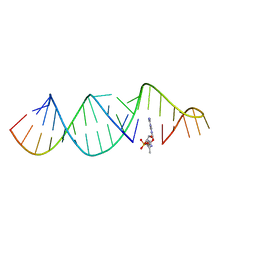 | | RNA hairpin structure containing tetraloop/receptor motif, complexed with 2-MeImpG analogue | | Descriptor: | 5'-O-[(S)-hydroxy(4-methyl-1H-imidazol-5-yl)phosphoryl]guanosine, RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*CP*CP*CP*GP*AP*AP*AP*GP*G)-3'), RNA (5'-R(P*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2017-03-31 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Rationale for the Enhanced Catalysis of Nonenzymatic RNA Primer Extension by a Downstream Oligonucleotide.
J. Am. Chem. Soc., 140, 2018
|
|
8TAR
 
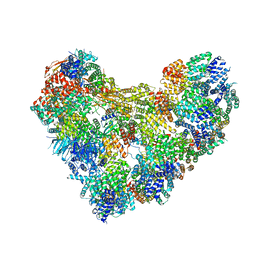 | | APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
8TAU
 
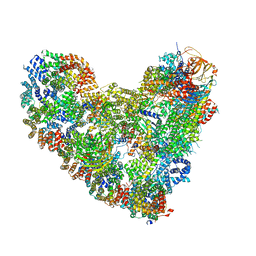 | | APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
5VBT
 
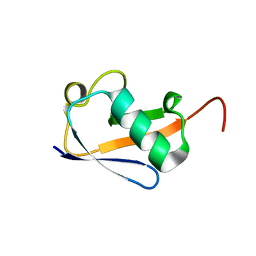 | | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor | | Descriptor: | UBH04 | | Authors: | DONG, A, DONG, X, LIU, L, GUO, Y, LI, Y, ZHANG, W, WALKER, J.R, SIDHU, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of a highly specific and potent USP7 ubiquitin variant inhibitor
to be published
|
|
7C8U
 
 | | The crystal structure of COVID-19 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Luan, X, Shang, W, Wang, Y, Yin, W, Jiang, Y, Feng, S, Wang, Y, Liu, M, Zhou, R, Zhang, Z, Wang, F, Cheng, W, Gao, M, Wang, H, Wu, W, Tian, R, Tian, Z, Jin, Y, Jiang, H.W, Zhang, L, Xu, H.E, Zhang, S. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of COVID-19 main protease in complex with GC376
To Be Published
|
|
1THD
 
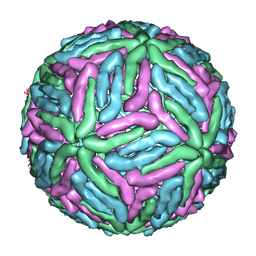 | | COMPLEX ORGANIZATION OF DENGUE VIRUS E PROTEIN AS REVEALED BY 9.5 ANGSTROM CRYO-EM RECONSTRUCTION | | Descriptor: | Major envelope protein E | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-06-01 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
4I5M
 
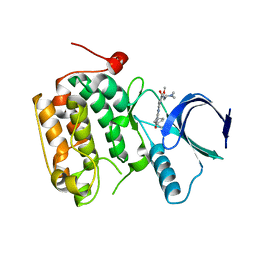 | | Selective & Brain-Permeable Polo-like Kinase-2 (Plk-2) Inhibitors that Reduce -Synuclein Phosphorylation in Rat Brain | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK2 | | Authors: | Aubele, D.L, Hom, R.K, Adler, M, Galemmo Jr, R.A, Bowers, S, Truong, A.P, Pan, H, Beroza, P, Neitz, R.J, Yao, N, Lin, M, Tonn, G, Zhang, H, Bova, M.P, Ren, Z, Tam, D, Ruslim, L, Baker, J, Diep, L, Fitzgerald, K, Hoffman, J, Motter, R, Fauss, D, Tanaka, P, Dappen, M, Jagodzinski, J, Chan, W, Konradi, A.W, Latimer, L, Zhu, Y.L, Artis, D.R, Sham, H.L, Anderson, J.P, Bergeron, M. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain.
Chemmedchem, 8, 2013
|
|
3KQ8
 
 | |
5C7M
 
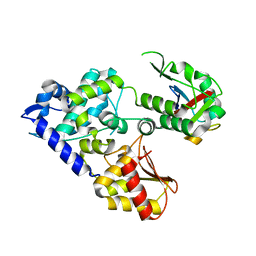 | | CRYSTAL STRUCTURE OF E3 LIGASE ITCH WITH A UB VARIANT | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Polyubiquitin-C | | Authors: | Walker, J.R, Hu, J, Dong, A, Wernimont, A, Zhang, W, Sidhu, S, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-24 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes.
Mol.Cell, 62, 2016
|
|
1TGE
 
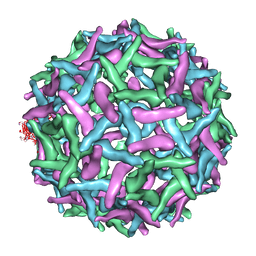 | | The structure of immature Dengue virus at 12.5 angstrom | | Descriptor: | envelope glycoprotein | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-05-28 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein.
Structure, 12, 2004
|
|
4RI2
 
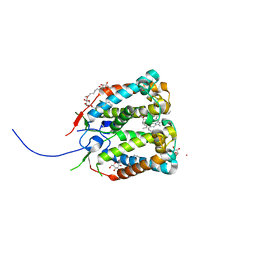 | | Crystal structure of the photoprotective protein PsbS from spinach | | Descriptor: | CHLOROPHYLL A, MERCURY (II) ION, Photosystem II 22 kDa protein, ... | | Authors: | Fan, M, Li, M, Chang, W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of the PsbS protein essential for photoprotection in plants.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RI3
 
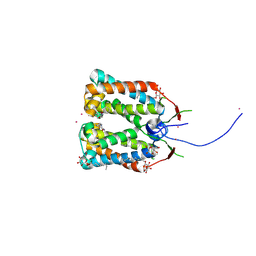 | | Crystal structure of DCCD-modified PsbS from spinach | | Descriptor: | DICYCLOHEXYLUREA, MERCURY (II) ION, Photosystem II 22 kDa protein, ... | | Authors: | Fan, M, Li, M, Chang, W. | | Deposit date: | 2014-10-05 | | Release date: | 2015-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the PsbS protein essential for photoprotection in plants.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1SWV
 
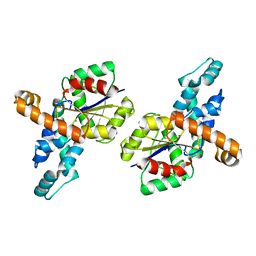 | | Crystal structure of the D12A mutant of phosphonoacetaldehyde hydrolase complexed with magnesium | | Descriptor: | MAGNESIUM ION, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
1RQL
 
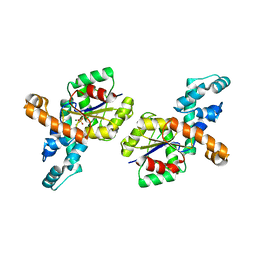 | | Crystal Structure of Phosponoacetaldehyde Hydrolase Complexed with Magnesium and the Inhibitor Vinyl Sulfonate | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase, VINYLSULPHONIC ACID | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
3LTU
 
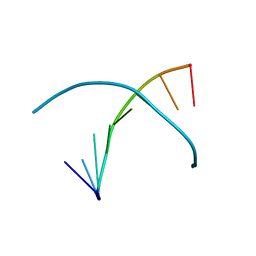 | | 5-SeMe-dU containing DNA 8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T5S)P*AP*CP*AP*C)-3' | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
3LTR
 
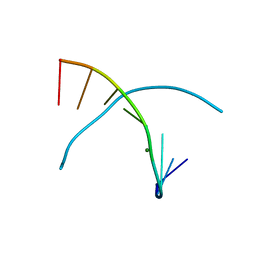 | | 5-OMe-dU containing DNA 8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T5O)P*AP*CP*AP*C)-3', MAGNESIUM ION | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen bond formation between the naturally modified nucleobase and phosphate backbone.
Nucleic Acids Res., 40, 2012
|
|
6A9S
 
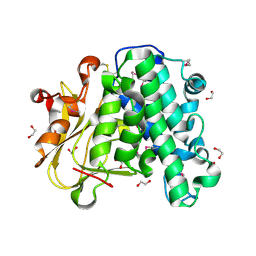 | | The crystal structure of vaccinia virus A26 (residues 1-397) | | Descriptor: | 1,2-ETHANEDIOL, Protein A26 | | Authors: | Wang, H.C, Ko, T.Z, Luo, Y.C, Liao, Y.T, Chang, W. | | Deposit date: | 2018-07-16 | | Release date: | 2019-06-12 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Vaccinia viral A26 protein is a fusion suppressor of mature virus and triggers membrane fusion through conformational change at low pH.
Plos Pathog., 15, 2019
|
|
8INI
 
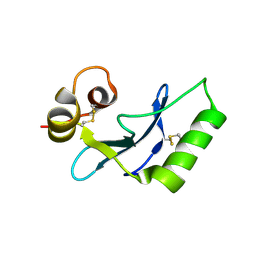 | | vaccinia H2 protein without the transmembrane region | | Descriptor: | TETRAETHYLENE GLYCOL, vaccinia h2 protein | | Authors: | Liu, C.Y, Ko, T.P, Wang, H.C, Chang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional analyses of viral H2 protein of the vaccinia virus entry fusion complex.
J.Virol., 97, 2023
|
|
