5XSD
 
 | | XylFII-LytSN complex mutant - D103A | | 分子名称: | Periplasmic binding protein/LacI transcriptional regulator, Signal transduction histidine kinase, LytS | | 著者 | Li, J.X, Wang, C.Y, Zhang, P. | | 登録日 | 2017-06-13 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6KZ8
 
 | | Crystal structure of plant Phospholipase D alpha complex with phosphatidic acid | | 分子名称: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, CALCIUM ION, Phospholipase D alpha 1 | | 著者 | Li, J.X, Yu, F, Zhang, P. | | 登録日 | 2019-09-23 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.291 Å) | | 主引用文献 | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
7BHS
 
 | | Crystal structure of MAT2a with quinazoline fragment 2 bound in the allosteric site | | 分子名称: | 6-chloranyl-2-methoxy-4-phenyl-quinazoline, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHU
 
 | | Crystal structure of MAT2a with elaborated fragment 26 bound in the allosteric site | | 分子名称: | 1,2-ETHANEDIOL, 7-chloranyl-4-(dimethylamino)-1-(2-hydroxyethyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, ... | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHW
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 29) | | 分子名称: | 7-chloranyl-4-(dimethylamino)-1-(3-methylphenyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHR
 
 | | Crystal structure of MAT2a with triazinone fragment 1 bound in the allosteric site | | 分子名称: | 4-(dimethylamino)-6-ethoxy-1~{H}-1,3,5-triazin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHV
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor and in vivo tool compound 28 | | 分子名称: | 7-chloranyl-4-(dimethylamino)-1-phenyl-quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHT
 
 | | Crystal structure of MAT2a with quinazolinone fragment 5 bound in the allosteric site | | 分子名称: | 7-chloranyl-4-(dimethylamino)-1~{H}-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.052 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHX
 
 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 31) | | 分子名称: | 7-chloranyl-4-(dimethylamino)-1-pyridin-3-yl-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | 登録日 | 2021-01-11 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
3JA6
 
 | | Cryo-electron Tomography and All-atom Molecular Dynamics Simulations Reveal a Novel Kinase Conformational Switch in Bacterial Chemotaxis Signaling | | 分子名称: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein 2 | | 著者 | Cassidy, C.K, Himes, B.A, Alvarez, F.J, Ma, J, Zhao, G, Perilla, J.R, Schulten, K, Zhang, P. | | 登録日 | 2015-04-21 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (12.7 Å) | | 主引用文献 | CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling.
Elife, 4, 2015
|
|
6KZ9
 
 | |
1LQG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | 分子名称: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | 著者 | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | 登録日 | 2002-05-10 | | 公開日 | 2002-11-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3J4F
 
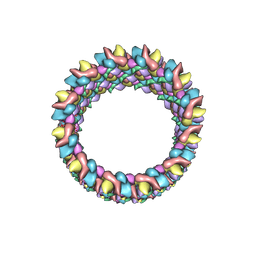 | | Structure of HIV-1 capsid protein by cryo-EM | | 分子名称: | capsid protein | | 著者 | Zhao, G, Perilla, J.R, Meng, X, Schulten, K, Zhang, P. | | 登録日 | 2013-07-11 | | 公開日 | 2013-07-24 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (8.6 Å) | | 主引用文献 | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | 分子名称: | capsid protein | | 著者 | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | 登録日 | 2013-02-23 | | 公開日 | 2013-05-29 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (8.6 Å) | | 主引用文献 | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
6LZ3
 
 | |
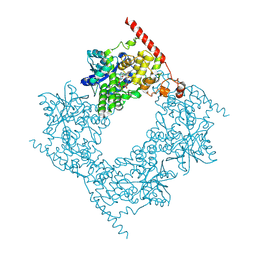
6LZ7
 
 | |
5XSS
 
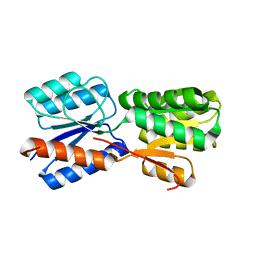 | | XylFII molecule | | 分子名称: | Periplasmic binding protein/LacI transcriptional regulator, beta-D-xylopyranose | | 著者 | Li, J.X, Wang, C.Y, Zhang, P. | | 登録日 | 2017-06-15 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.093 Å) | | 主引用文献 | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3IWP
 
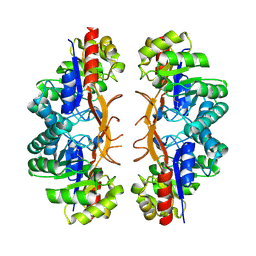 | |
3J3Y
 
 | |
5Z6P
 
 | | The crystal structure of an agarase, AgWH50C | | 分子名称: | B-agarase | | 著者 | Mao, X, Zhou, J, Zhang, P, Zhang, L, Zhang, J, Li, Y. | | 登録日 | 2018-01-24 | | 公開日 | 2019-01-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.061 Å) | | 主引用文献 | Structure-based design of agarase AgWH50C from Agarivorans gilvus WH0801 to enhance thermostability.
Appl. Microbiol. Biotechnol., 103, 2019
|
|
3HFN
 
 | |
6K9L
 
 | | 4.27 Angstrom resolution cryo-EM structure of human dimeric ATM kinase | | 分子名称: | Serine-protein kinase ATM | | 著者 | Xiao, J, Liu, M, Qi, Y, Chaban, Y, Gao, C, Tian, Y, Yu, Z, Li, J, Zhang, P, Xu, Y. | | 登録日 | 2019-06-16 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.27 Å) | | 主引用文献 | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
6QLZ
 
 | | IDOL F3ab subdomain | | 分子名称: | E3 ubiquitin-protein ligase MYLIP | | 著者 | Martinelli, L, Johansson, P, Wan, P.T, Gunnarsson, J, Guo, H, Boyd, H. | | 登録日 | 2019-02-01 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.343 Å) | | 主引用文献 | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
6KVL
 
 | | Crystal structure of UDP-RebB-SrUGT76G1 | | 分子名称: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | 著者 | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | 登録日 | 2019-09-04 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KVI
 
 | | Crystal structure of UDP-SrUGT76G1 | | 分子名称: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | 著者 | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | 登録日 | 2019-09-04 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
