6KMM
 
 | | Crystal Structure of HEPES bound Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dhankhar, P, Dalal, V, Mahto, J.K, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Characterization of dye-decolorizing peroxidase from Bacillus subtilis.
Arch.Biochem.Biophys., 693, 2020
|
|
6KMN
 
 | | Crystal Structure of Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Deferrochelatase/peroxidase EfeB, ... | | Authors: | Dhankhar, P, Dalal, V, Mahto, J.K, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Characterization of dye-decolorizing peroxidase from Bacillus subtilis.
Arch.Biochem.Biophys., 693, 2020
|
|
2BNG
 
 | | Structure of an M.tuberculosis LEH-like epoxide hydrolase | | Descriptor: | CALCIUM ION, MB2760 | | Authors: | Johansson, P, Arand, M, Unge, T, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2005-03-24 | | Release date: | 2005-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an Atypical Epoxide Hydrolase from Mycobacterium Tuberculosis Gives Insights Into its Function.
J.Mol.Biol., 351, 2005
|
|
2C2I
 
 | | Structure and function of Rv0130, a conserved hypothetical protein from M.tuberculosis | | Descriptor: | RV0130 | | Authors: | Johansson, P, Castell, A, Jones, T.A, Backbro, K. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of Rv0130, a Conserved Hypothetical Protein from Mycobacterium Tuberculosis.
Protein Sci., 15, 2006
|
|
4QDM
 
 | | Crystal structure of N-terminal mutant (V1L) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Alkaline thermostable endoxylanase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2014-05-14 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | Structural insights into N-terminal to C-terminal interactions and implications for thermostability of a (beta/alpha)8-triosephosphate isomerase barrel enzyme
Febs J., 282, 2015
|
|
2MTZ
 
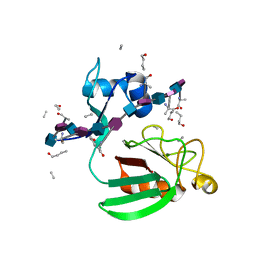 | | Haddock model of Bacillus subtilis L,D-transpeptidase in complex with a peptidoglycan hexamuropeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Putative L,D-transpeptidase YkuD, intact bacterial peptidoglycan | | Authors: | Schanda, P, Triboulet, S, Laguri, C, Bougault, C, Ayala, I, Callon, M, Arthur, M, Simorre, J. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic model of a cell-wall cross-linking enzyme in complex with an intact bacterial peptidoglycan.
J.Am.Chem.Soc., 136, 2014
|
|
1SB2
 
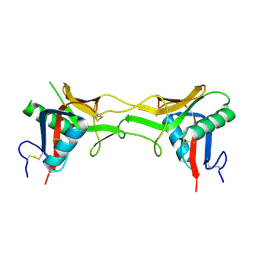 | | High resolution Structure determination of rhodocetin | | Descriptor: | Rhodocetin alpha subunit, Rhodocetin beta subunit | | Authors: | Paaventhan, P, Kong, C.G, Joseph, J.S, Chung, M.C.M, Kolatkar, P.R. | | Deposit date: | 2004-02-10 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of rhodocetin reveals noncovalently bound heterodimer interface
Protein Sci., 14, 2005
|
|
4QCE
 
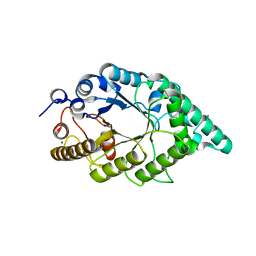 | | Crystal structure of recombinant alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Alkaline thermostable endoxylanase, MAGNESIUM ION, SODIUM ION | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2014-05-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural insights into N-terminal to C-terminal interactions and implications for thermostability of a (beta/alpha)8-triosephosphate isomerase barrel enzyme
Febs J., 282, 2015
|
|
4QCF
 
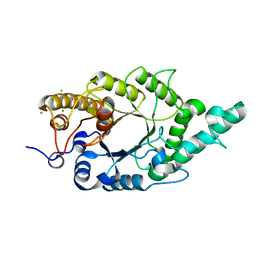 | | Crystal structure of N-terminal mutant (V1A) of an alkali thermostable GH10 xylanase from Bacillus sp. NG-27 | | Descriptor: | Alkaline thermostable endoxylanase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mahanta, P, Bhardwaj, A, Reddy, V.S, Ramakumar, S. | | Deposit date: | 2014-05-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insights into N-terminal to C-terminal interactions and implications for thermostability of a (beta/alpha)8-triosephosphate isomerase barrel enzyme
Febs J., 282, 2015
|
|
9IK2
 
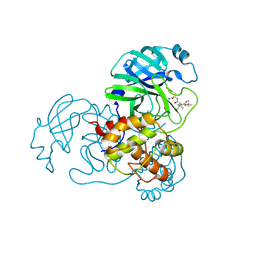 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H109 | | Descriptor: | 3C-like proteinase, tert-butyl N-[(2S)-1-[[(2S)-1-[[(2S)-1-azanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Feng, Y, Zheng, W.Y, Han, P, Fu, L.F, Qi, J.X. | | Deposit date: | 2024-06-26 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a small molecule inhibitor of SARS-CoV-2 main protease with potent in vitro and in vivo antiviral activities
To Be Published
|
|
3J3Q
 
 | |
6XF8
 
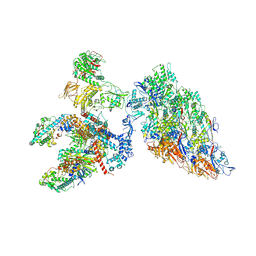 | | DLP 5 fold | | Descriptor: | Inner capsid protein lambda-1, Inner capsid protein sigma-2, Outer capsid protein mu-1, ... | | Authors: | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
6XF7
 
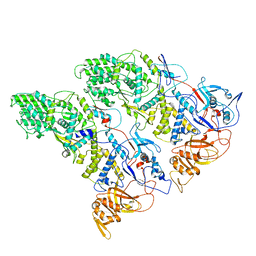 | | SLP | | Descriptor: | Lambda 1 protein | | Authors: | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
8Q71
 
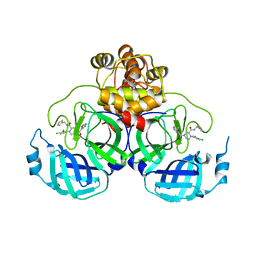 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GC-67 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-(4-methoxypyridin-3-yl)carbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Weisse, R.H, Useini, A, Gao, S, Song, L, Liu, Z, Zhan, P. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Trisubstituted Piperazine Derivatives as Noncovalent Severe Acute Respiratory Syndrome Coronavirus 2 Main Protease Inhibitors with Improved Antiviral Activity and Favorable Druggability.
J.Med.Chem., 66, 2023
|
|
7NH9
 
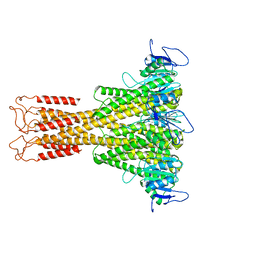 | |
8C9M
 
 | | HERV-K Gag immature lattice | | Descriptor: | Gag protein | | Authors: | Krebs, A.-S, Liu, H.-F, Zhou, Y, Rey, J.S, Levintov, L, Perilla, J.R, Bartesaghi, A, Zhang, P. | | Deposit date: | 2023-01-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and conservation of an immature human endogenous retrovirus.
Biorxiv, 2023
|
|
5X41
 
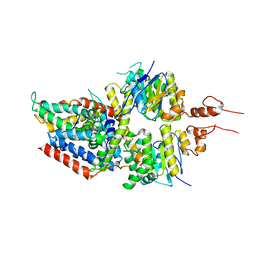 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X40
 
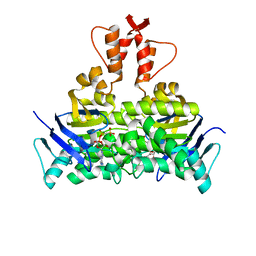 | | Structure of a CbiO dimer bound with AMPPCP | | Descriptor: | Cobalt ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5XN6
 
 | | Heterodimer crystal structure of geranylgeranyl diphosphate synthases 1 with GGPPS Recruiting Protein(OsGRP) from Oryza sativa | | Descriptor: | Os02g0668100 protein, Os07g0580900 protein | | Authors: | Wang, C, Zhou, F, Lu, S, Zhang, P. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.598 Å) | | Cite: | A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8C5V
 
 | |
7ZBT
 
 | | Subtomogram averaging of Rubisco from native Halothiobacillus carboxysomes | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small subunit | | Authors: | Ni, T, Zhu, Y, Yu, X, Sun, Y, Liu, L, Zhang, P. | | Deposit date: | 2022-03-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and assembly of cargo Rubisco in two native alpha-carboxysomes.
Nat Commun, 13, 2022
|
|
7ZC1
 
 | | Subtomogram averaging of Rubisco from Cyanobium carboxysome | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase, small subunit | | Authors: | Ni, T, Zhu, Y, Seaton-Burn, W, Zhang, P. | | Deposit date: | 2022-03-25 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and assembly of cargo Rubisco in two native alpha-carboxysomes.
Nat Commun, 13, 2022
|
|
7YZY
 
 | | pMMO structure from native membranes by cryoET and STA | | Descriptor: | Methane monooxygenase subunit C2, Particulate methane monooxygenase alpha subunit, Particulate methane monooxygenase beta subunit | | Authors: | Zhu, Y, Ni, T, Zhang, P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and activity of particulate methane monooxygenase arrays in methanotrophs.
Nat Commun, 13, 2022
|
|
1KQ1
 
 | | 1.55 A Crystal structure of the pleiotropic translational regulator, Hfq | | Descriptor: | ACETIC ACID, Host Factor for Q beta | | Authors: | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | Deposit date: | 2002-01-03 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
1KQ2
 
 | | Crystal Structure of an Hfq-RNA Complex | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*G)-3', Host factor for Q beta | | Authors: | Schumacher, M.A, Pearson, R.F, Moller, T, Valentin-Hansen, P, Brennan, R.G. | | Deposit date: | 2002-01-03 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein.
EMBO J., 21, 2002
|
|
