5HIL
 
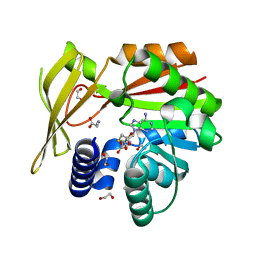 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with S-adenosylhomocysteine and sarcosine | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity
Sci Rep, 6, 2016
|
|
1PVZ
 
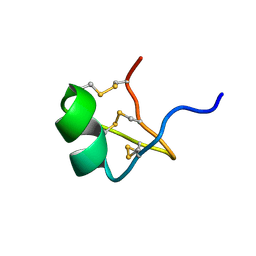 | | Solution Structure of BmP07, A Novel Potassium Channel Blocker from Scorpion Buthus martensi Karsch, 15 structures | | Descriptor: | K+ toxin-like peptide | | Authors: | Wu, H, Zhang, N, Wang, Y, Zhang, Q, Ou, L, Li, M, Hu, G. | | Deposit date: | 2003-06-29 | | Release date: | 2004-05-18 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK2, a new potassium channel blocker from the venom of chinese scorpion Buthus martensi Karsch
PROTEINS, 55, 2004
|
|
8TA8
 
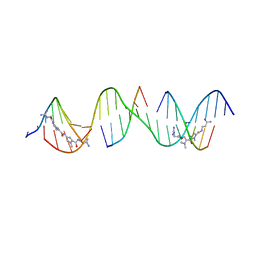 | | Sequence specific (AATT) orientation of netropsin molecules at two unique minor groove binding sites within a self-assembled 3D DNA lattice (4x6) | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*AP*TP*TP*CP*CP*TP*GP*AP*CP*GP*GP*AP*AP*AP*TP*TP*A*(NT)*(NT))-3'), DNA (5'-D(*TP*CP*TP*AP*AP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-26 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TAM
 
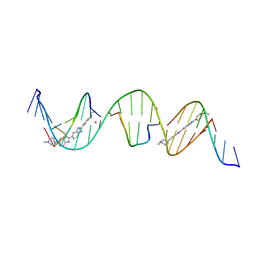 | | Sequence specific (AATT) orientation of Hoechst molecules at two unique minor groove binding sites within a self-assembled 3D DNA lattice (4x6) | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, COBALT (II) ION, DNA (5'-D(*GP*AP*GP*AP*AP*TP*TP*CP*CP*TP*GP*AP*CP*GP*GP*AP*AP*AP*TP*TP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-27 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TBO
 
 | | Sequence specific (AATT) orientation of netropsin molecules at a unique minor groove binding site (position1) within a self-assembled 3D DNA lattice (4x5) | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*CP*AP*AP*TP*TP*GP*CP*TP*GP*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TB4
 
 | | Sequence specific (AATT) orientation of DAPI molecules at a unique minor groove binding site (position2) within a self-assembled 3D DNA lattice (4x5) | | Descriptor: | 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*TP*TP*GP*T*(DAP))-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-28 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TB3
 
 | | Sequence specific (AATT) orientation of DAPI molecules at a unique minor groove binding site (position1) within a self-assembled 3D DNA lattice (4x5) | | Descriptor: | 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, DNA (5'-D(*GP*AP*CP*AP*AP*TP*TP*GP*CP*TP*GP*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-28 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TDT
 
 | | Sequence specific (AATT) orientation of DAPI molecules at a unique minor groove binding site (position2) within a self-assembled 3D DNA lattice (4x6) | | Descriptor: | 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*GP*AP*AP*AP*TP*TP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-07-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.007 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TAQ
 
 | | Sequence specific (AATT) orientation of Hoechst molecules at a unique minor groove binding site (position2) within a self-assembled 3D DNA lattice (4x6) | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*GP*AP*AP*AP*TP*TP*A*(HT1))-3'), DNA (5'-D(*TP*CP*TP*AP*AP*TP*TP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-27 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TBD
 
 | | Sequence specific (AATT) orientation of Hoechst molecules at a unique minor groove binding site (position2) within a self-assembled 3D DNA lattice (4x5) | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*AP*TP*TP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-28 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8T7B
 
 | | Sequence specific (AATT) orientation of netropsin molecules at a unique minor groove binding site (position2) within a self-assembled 3D DNA lattice (4x5) | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*TP*TP*G)-3'), DNA (5'-D(P*AP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TC2
 
 | | Sequence specific (AATT) orientation of netropsin molecules at two unique minor groove binding sites within a self-assembled 3D DNA lattice (4x5) | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*TP*TP*G)-3'), DNA (5'-D(P*AP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8T80
 
 | | Sequence specific (AATT) orientation of Hoechst molecules at two unique minor groove binding sites within a self-assembled 3D DNA lattice (4x5) | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*GP*AP*GP*AP*AP*TP*TP*CP*CP*TP*GP*AP*CP*GP*AP*CP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*TP*TP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-21 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TA9
 
 | | Sequence specific (AATT) orientation of DAPI molecules at a unique minor groove binding site (position1) within a self-assembled 3D DNA lattice (4x6) | | Descriptor: | 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, COBALT (II) ION, DNA (5'-D(*GP*AP*GP*AP*AP*TP*TP*CP*CP*TP*GP*AP*CP*GP*GP*AP*AP*CP*TP*CP*A*(DAP))-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-27 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TAP
 
 | | Sequence specific (AATT) orientation of netropsin molecules at a unique minor groove binding site (position2) within a self-assembled 3D DNA lattice (4x6) | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*GP*AP*CP*GP*GP*AP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*CP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-27 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TB8
 
 | | Sequence specific (AATT) orientation of Hoechst molecules at a unique minor groove binding site (position1) within a self-assembled 3D DNA lattice (4x5) | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*GP*AP*CP*AP*AP*TP*TP*GP*CP*TP*GP*AP*CP*GP*AP*CP*AP*CP*TP*CP*A*(HT1))-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-28 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
8TAJ
 
 | | Sequence specific (AATT) orientation of netropsin molecules at a unique minor groove binding site (position1) within a self-assembled 3D DNA lattice (4x6) | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*AP*AP*TP*TP*CP*CP*TP*GP*AP*CP*GP*GP*AP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*T)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2023-06-27 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Site-Specific Arrangement and Structure Determination of Minor Groove Binding Molecules in Self-Assembled Three-Dimensional DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
5J7D
 
 | | Computationally Designed Thioredoxin dF106 | | Descriptor: | COPPER (II) ION, Designed Thioredoxin dF106 | | Authors: | Horowitz, S, Johansen, N, Olsen, J.G, Winther, J.R. | | Deposit date: | 2016-04-06 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational Redesign of Thioredoxin Is Hypersensitive toward Minor Conformational Changes in the Backbone Template.
J.Mol.Biol., 428, 2016
|
|
5GWK
 
 | | Human topoisomerase IIalpha in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Wang, Y.R, Wu, C.C, Chan, N.L. | | Deposit date: | 2016-09-12 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.152 Å) | | Cite: | Producing irreversible topoisomerase II-mediated DNA breaks by site-specific Pt(II)-methionine coordination chemistry
Nucleic Acids Res., 45, 2017
|
|
3Q3J
 
 | | Crystal structure of plexin A2 RBD in complex with Rnd1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-A2, ... | | Authors: | Wang, H, Tempel, W, Tong, Y, Guan, X, Shen, L, Buren, L, Zhang, N, Wernimont, A.K, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-21 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Crystal structure of plexin A2 RBD in complex with Rnd1
to be published
|
|
2MUX
 
 | |
3RBT
 
 | | Crystal structure of glutathione S-transferase Omega 3 from the silkworm Bombyx mori | | Descriptor: | GLYCEROL, Glutathione transferase o1 | | Authors: | Chen, B.-Y, Ma, X.-X, Tan, X, Yang, J.-P, Zhang, N.-N, Li, W.-F, Chen, Y, Xia, Q, Zhou, C.-Z. | | Deposit date: | 2011-03-29 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided activity restoration of the silkworm glutathione transferase Omega GSTO3-3
J.Mol.Biol., 412, 2011
|
|
3WJA
 
 | | The crystal structure of human cytosolic NADP(+)-dependent malic enzyme in apo form | | Descriptor: | NADP-dependent malic enzyme | | Authors: | Li, S.-Y, Chen, M.-C, Yang, P.-C, Chan, N.-L, Liu, J.-H, Hung, H.-C. | | Deposit date: | 2013-10-08 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | Structural characteristics of the nonallosteric human cytosolic malic enzyme.
Biochim.Biophys.Acta, 1844, 2014
|
|
3O16
 
 | | Crystal Structure of Bacillus subtilis Thiamin Phosphate Synthase K159A | | Descriptor: | Thiamine-phosphate pyrophosphorylase | | Authors: | McCulloch, K.M, Hanes, J.W, Abdelwahed, S, Mahanta, N, Hazra, A, Ishida, K, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-07-20 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Kinetic Characterization of Bacillus subtilis Thiamin
Phosphate Synthase with a Carboxylated Thiazole Phosphate
to be published
|
|
4ZGY
 
 | |
