7C3E
 
 | | Crystal structure of R97A/R150A/R203A mutant of AofleA from Arthrobotrys oligospora | | Descriptor: | AofleA, GLYCEROL, SULFATE ION | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7C3C
 
 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with D-manose | | Descriptor: | AofleA, GLYCEROL, alpha-D-mannopyranose, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7C37
 
 | | Crystal structure of AofleA from Arthrobotrys oligospora | | Descriptor: | AofleA, BICINE | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
5WBX
 
 | |
5WC5
 
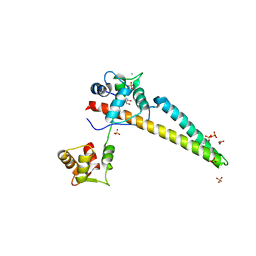 | |
7CHA
 
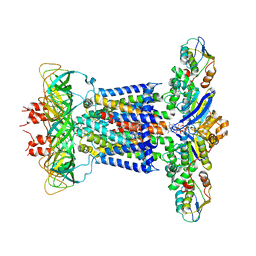 | | Cryo-EM structure of P.aeruginosa MlaFEBD with AMPPNP | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CH6
 
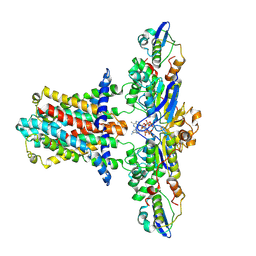 | | Cryo-EM structure of E.coli MlaFEB with AMPPNP | | Descriptor: | Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
5J6P
 
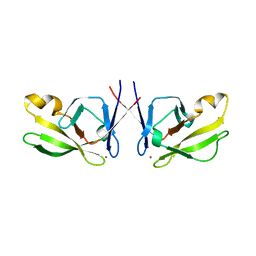 | | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe | | Descriptor: | Kinetochore protein mis18, ZINC ION | | Authors: | Wang, C, Shao, C, Zhang, M, Zhang, X, Zang, J. | | Deposit date: | 2016-04-05 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe
To Be Published
|
|
1B8Q
 
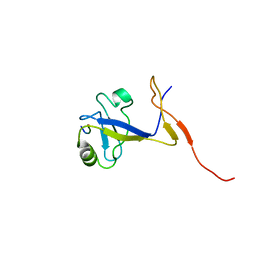 | | SOLUTION STRUCTURE OF THE EXTENDED NEURONAL NITRIC OXIDE SYNTHASE PDZ DOMAIN COMPLEXED WITH AN ASSOCIATED PEPTIDE | | Descriptor: | PROTEIN (HEPTAPEPTIDE), PROTEIN (NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Tochio, H, Zhang, Q, Mandal, P, Li, M, Zhang, M. | | Deposit date: | 1999-02-01 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide.
Nat.Struct.Biol., 6, 1999
|
|
6MG0
 
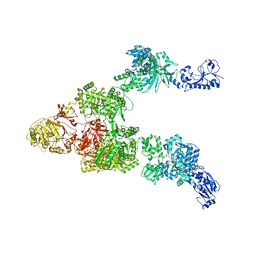 | | Crystal structure of a 5-domain construct of LgrA in the thiolation state | | Descriptor: | 5'-({[(2R,3R)-3-amino-2-{[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-{[oxido(oxo)phosphonio]oxy}butanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-4-methylpentyl]sulfonyl}amino)-5'-deoxyadenosine, Linear gramicidin synthase subunit A | | Authors: | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MFW
 
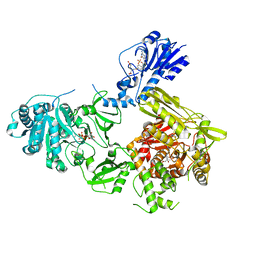 | | Crystal structure of a 4-domain construct of LgrA in the substrate donation state | | Descriptor: | (2~{R})-~{N}-[3-[2-[[(2~{S})-2-formamido-3-methyl-butanoyl]amino]ethylamino]-3-oxidanylidene-propyl]-3,3-dimethyl-2-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-butanamide, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Linear gramicidin synthase subunit A, ... | | Authors: | Reimer, J.M, Eivaskhani, M, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MFY
 
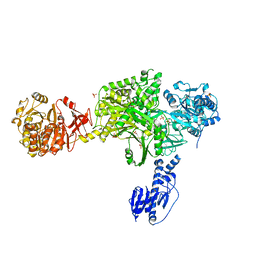 | | Crystal structure of a 5-domain construct of LgrA in the substrate donation state | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Linear gramicidin synthase subunit A, PHOSPHATE ION | | Authors: | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MFX
 
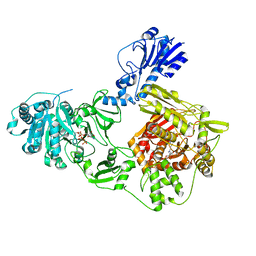 | | Crystal structure of a 4-domain construct of a mutant of LgrA in the substrate donation state | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Linear gramicidin synthase subunit A, N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Reimer, J.M, Eivaskhani, M, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6MFZ
 
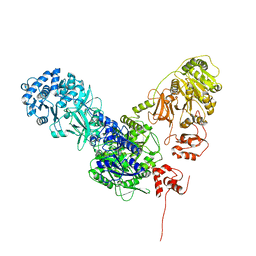 | | Crystal structure of dimodular LgrA in a condensation state | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Linear gramicidin synthase subunit A | | Authors: | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
7SWL
 
 | | CryoEM structure of the N-terminal-deleted Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
7T0V
 
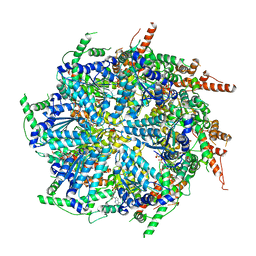 | | CryoEM structure of the crosslinked Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-30 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
4RZ0
 
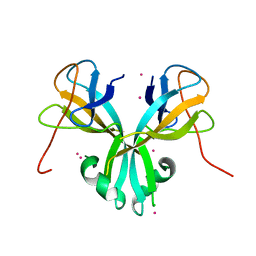 | | Crystal Structure of Plasmodium falciparum putative histone methyltransferase PFL0690c | | Descriptor: | PFL0690c, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-17 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Crystal Structure of Plasmodium falciparum putative histone methyltransferase PFL0690c
To be Published
|
|
4YL8
 
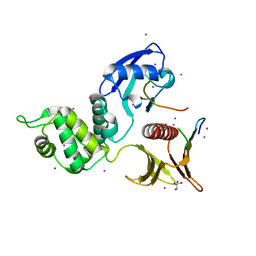 | | Crystal structure of the Crumbs/Moesin complex | | Descriptor: | GLYCEROL, IODIDE ION, Moesin, ... | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for the Phosphorylation-regulated Interaction between the Cytoplasmic Tail of Cell Polarity Protein Crumbs and the Actin-binding Protein Moesin
J.Biol.Chem., 290, 2015
|
|
4YTL
 
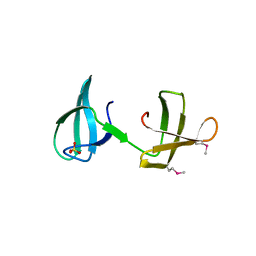 | | Structure of the KOW2-KOW3 Domain of Transcription Elongation Factor Spt5. | | Descriptor: | GLYCEROL, SULFATE ION, Transcription elongation factor SPT5 | | Authors: | Meyer, P.A, Li, S, ZHang, M, Yamada, K, Takagi, Y, Hartzog, G.A, Fu, J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5.
Mol.Cell.Biol., 35, 2015
|
|
4DC2
 
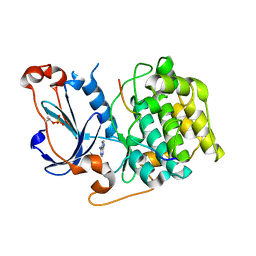 | | Structure of PKC in Complex with a Substrate Peptide from Par-3 | | Descriptor: | ADENINE, Partitioning defective 3 homolog, Protein kinase C iota type | | Authors: | Shang, Y, Wang, C, Yu, J, Zhang, M. | | Deposit date: | 2012-01-17 | | Release date: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKC iota in complex with a substrate peptide from Par-3
Structure, 20, 2012
|
|
4ZRK
 
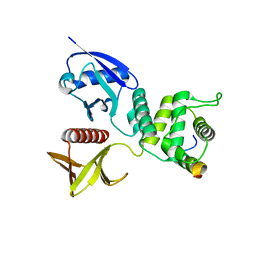 | | Merlin-FERM and Lats1 complex | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS1 | | Authors: | Lin, Z, Li, Y, Wei, Z, Zhang, M. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
6CZQ
 
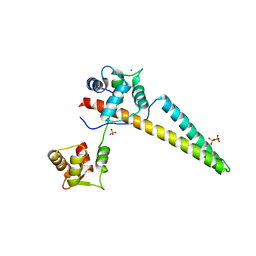 | |
1M5Z
 
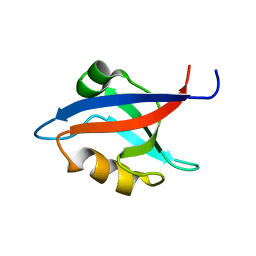 | | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area | | Descriptor: | AMPA receptor interacting protein | | Authors: | Feng, W, Fan, J, Jiang, M, Shi, Y, Zhang, M. | | Deposit date: | 2002-07-11 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The PDZ7 of Glutamate Receptor Interacting Protein Binds to its Target via a Novel Hydrophobic Surface Area
J.Biol.Chem., 277, 2002
|
|
4YTK
 
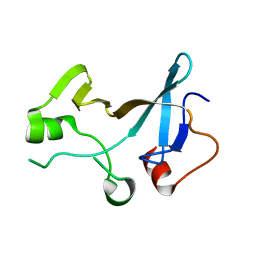 | | Structure of the KOW1-Linker1 domain of Transcription Elongation Factor Spt5 | | Descriptor: | Transcription elongation factor SPT5 | | Authors: | Meyer, P.A, Li, S, Zhang, M, Yamada, K, Takagi, Y, Hartzog, G.A, Fu, J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.0904 Å) | | Cite: | Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5.
Mol.Cell.Biol., 35, 2015
|
|
7CBO
 
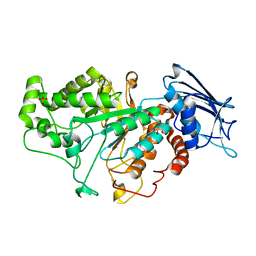 | | Crystal structure of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL, ... | | Authors: | Xu, W, Wang, M, Zhang, M. | | Deposit date: | 2020-06-13 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and biochemical analyses of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila involved in mucin degradation.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
