3X0G
 
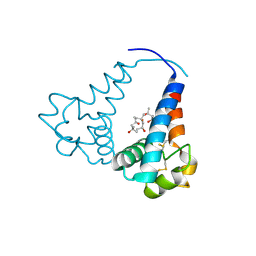 | |
2Z8Q
 
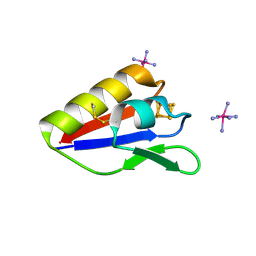 | |
2IB6
 
 | | Structural characterization of a blue chromoprotein and its yellow mutant from the sea anemone cnidopus japonicus | | Descriptor: | PHOSPHATE ION, Yellow mutant chromo protein | | Authors: | Chan, M.C.Y, Bosanac, I, Ho, D, Prive, G, Ikura, M. | | Deposit date: | 2006-09-10 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of a Blue Chromoprotein and Its Yellow Mutant from the Sea Anemone Cnidopus Japonicus
J.Biol.Chem., 281, 2006
|
|
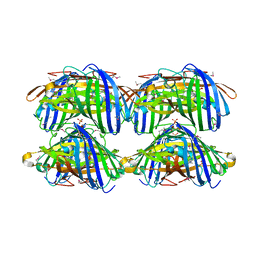
3D4S
 
 | | Cholesterol bound form of human beta2 adrenergic receptor. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(tert-butylamino)-3-[(4-morpholin-4-yl-1,2,5-thiadiazol-3-yl)oxy]propan-2-ol, Beta-2 adrenergic receptor/T4-lysozyme chimera, ... | | Authors: | Hanson, M.A, Cherezov, V, Roth, C.B, Griffith, M.T, Jaakola, V.-P, Chien, E.Y.T, Velasquez, J, Kuhn, P, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor.
Structure, 16, 2008
|
|
1W7W
 
 | |
2IB5
 
 | |
3F71
 
 | |
7L7Y
 
 | | x-ray structure of the N-acetyltransferase Pcryo_0637 from psychrobacter cryohalolentis in the presence of UDP and acetyl-conezyme A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
7L82
 
 | | x-ray structure of the psychrobacter cryohalolentis Pcryo_0637 N-acetyltransferase in the presene of its reaction tetrahedral intermediate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Putative acetyl transferase protein, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
7L81
 
 | | x-ray structure of the psychrobacter cryohalolentis N-acetyltransferase Pcryo_0637 in the presence of coenzyme A and | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
7L7X
 
 | | X-ray structure of the Pcryo_0638 aminotransferase from Psychrobacter cryohalolentis | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DegT/DnrJ/EryC1/StrS aminotransferase, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
7L7Z
 
 | | x-ray structure of the N-acetyltransferase Pcryo_0637 from psychrobacter cryohalolentis in the presence of coenzyme A and UDP-di-N-acetyl-bacillosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Linehan, M.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-12-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Characterization of two enzymes from Psychrobacter cryohalolentis that are required for the biosynthesis of an unusual diacetamido-d-sugar.
J.Biol.Chem., 296, 2021
|
|
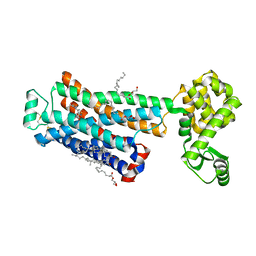
4DJC
 
 | | 1.35 A crystal structure of the NaV1.5 DIII-IV-Ca/CaM complex | | Descriptor: | CALCIUM ION, Calmodulin, ISOPROPYL ALCOHOL, ... | | Authors: | Sarhan, M.F, Tung, C.-C, Van Petegem, F, Ahern, C.A. | | Deposit date: | 2012-02-01 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystallographic basis for calcium regulation of sodium channels.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3EZG
 
 | |
7FDB
 
 | | CryoEM Structures of Reconstituted V-ATPase,State2 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDA
 
 | | CryoEM Structure of Reconstituted V-ATPase, state1 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDC
 
 | | CryoEM Structures of Reconstituted V-ATPase, state3 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDE
 
 | | CryoEM Structures of Reconstituted V-ATPase, Oxr1 bound V1 | | Descriptor: | Oxidation resistance protein 1, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
3B38
 
 | | Structure of A104V DJ-1 | | Descriptor: | 1,2-ETHANEDIOL, Protein DJ-1 | | Authors: | Lakshminarasimhan, M, Maldonado, M.T, Zhou, W, Fink, A.L, Wilson, M.A. | | Deposit date: | 2007-10-19 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Impact of Three Parkinsonism-Associated Missense Mutations on Human DJ-1.
Biochemistry, 47, 2008
|
|
3B36
 
 | | Structure of M26L DJ-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein DJ-1 | | Authors: | Lakshminarasimhan, M, Maldonado, M.T, Zhou, W, Fink, A.L, Wilson, M.A. | | Deposit date: | 2007-10-19 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Impact of Three Parkinsonism-Associated Missense Mutations on Human DJ-1.
Biochemistry, 47, 2008
|
|
3RKG
 
 | | Structural and Functional Characterization of the Yeast Mg2+ Channel Mrs2 | | Descriptor: | 1,2-ETHANEDIOL, Magnesium transporter MRS2, mitochondrial | | Authors: | Khan, M.B, Sponder, G, Sjoeblom, B, Svidova, S, Schweyen, R.J, Carugo, O, Djinovic-Carugo, K. | | Deposit date: | 2011-04-18 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural and functional characterization of the N-terminal domain of the yeast Mg(2+) channel Mrs2.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GQU
 
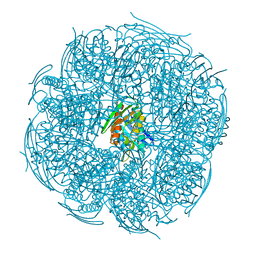 | | Crystal structure of HisB from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | Authors: | Ahangar, M.S, Vyas, R, Nasir, N, Biswal, B.K. | | Deposit date: | 2012-08-24 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of the native, substrate-
bound and inhibited forms of Mycobacterium tuberculosis imidazole glycerol phosphate dehydratase
Acta Crystallogr.,Sect.D, 2013
|
|
2JMM
 
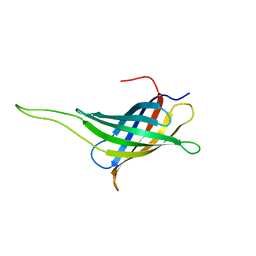 | | NMR solution structure of a minimal transmembrane beta-barrel platform protein | | Descriptor: | Outer membrane protein A | | Authors: | Johansson, M.U, Alioth, S, Hu, K, Walser, R, Koebnik, R, Pervushin, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-07-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | A minimal transmembrane beta-barrel platform protein studied by nuclear magnetic resonance
Biochemistry, 46, 2007
|
|
1PRB
 
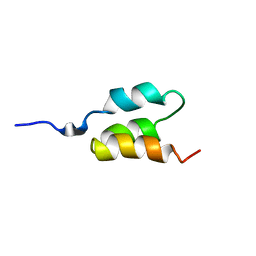 | | STRUCTURE OF AN ALBUMIN-BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN PAB | | Authors: | Johansson, M.U, De Chateau, M, Wikstrom, M, Forsen, S, Drakenberg, T, Bjorck, L. | | Deposit date: | 1997-01-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the albumin-binding GA module: a versatile bacterial protein domain.
J.Mol.Biol., 266, 1997
|
|
1AOR
 
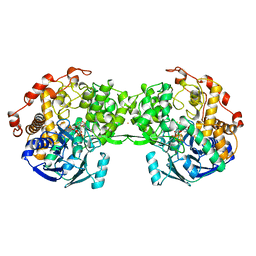 | | STRUCTURE OF A HYPERTHERMOPHILIC TUNGSTOPTERIN ENZYME, ALDEHYDE FERREDOXIN OXIDOREDUCTASE | | Descriptor: | ALDEHYDE FERREDOXIN OXIDOREDUCTASE, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Chan, M.K, Mukund, S, Kletzin, A, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1995-02-13 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase.
Science, 267, 1995
|
|
