7X2M
 
 | | Crystal structure of nanobody 1-2C7 with SARS-CoV-2 RBD | | Descriptor: | 1-2C7, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2J
 
 | | Crystal structure of nanobody Nb70 with SARS-CoV RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nb70, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
3ZWN
 
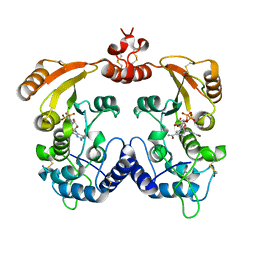 | | Crystal structure of Aplysia cyclase complexed with substrate NGD and product cGDPR | | Descriptor: | 3-(AMINOCARBONYL)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYD ROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2- YL]PYRIDINIUM, ADP-RIBOSYL CYCLASE, CYCLIC GUANOSINE DIPHOSPHATE-RIBOSE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
8JNP
 
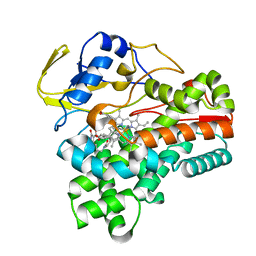 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with the substrate ikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,15R,16S,18Z,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.05,16.07,15.08,12]octacosa-1(2),3,13,18-tetraene-20,27,28-trione, Cytochrome P450 CftA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, P, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JNQ
 
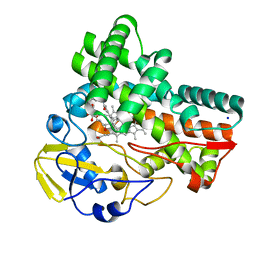 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with a substrate compound c | | Descriptor: | (1Z,3E,5E,7S,8R,10S,11R,13R,15R,16E,18E,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazatetracyclo[23.2.1.09,13.08,15]octacosa-1(2),3,5,16,18-pentaene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Jiang, P, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JFN
 
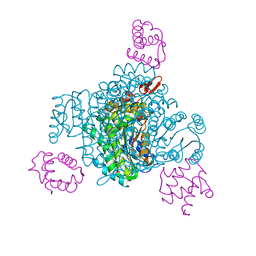 | |
8JF9
 
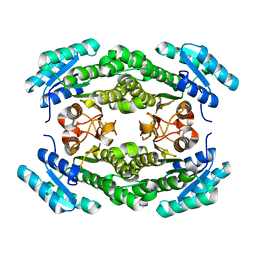 | |
5XJO
 
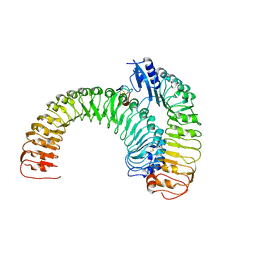 | | Plant receptor ERL1-TMM in complex with peptide EPF1 | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein EPIDERMAL PATTERNING FACTOR 1, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G, Zhang, L, Han, Z, Shpak, E.D, Yang, X. | | Deposit date: | 2017-05-03 | | Release date: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
7XSP
 
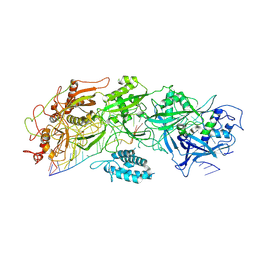 | | Structure of gRAMP-target RNA | | Descriptor: | RAMP superfamily protein, RNA (35-MER), RNA (5'-R(P*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | Authors: | Feng, Y, Zhang, L.X. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
8J6Z
 
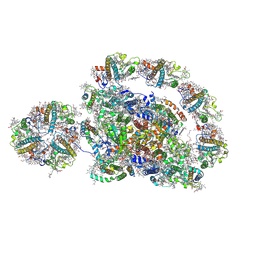 | | Cryo-EM structure of the Arabidopsis thaliana photosystem I(PSI-LHCII-ST2) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
8J7B
 
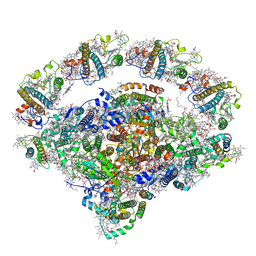 | | Coordinates of Cryo-EM structure of the Arabidopsis thaliana PSI in state 2 (PSI-ST2) | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | Deposit date: | 2023-04-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
8J7A
 
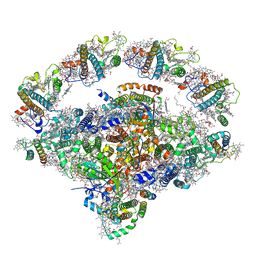 | | Coordinates of Cryo-EM structure of the Arabidopsis thaliana PSI in state 1 (PSI-ST1) | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | Deposit date: | 2023-04-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
1QCR
 
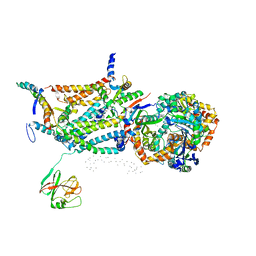 | | CRYSTAL STRUCTURE OF BOVINE MITOCHONDRIAL CYTOCHROME BC1 COMPLEX, ALPHA CARBON ATOMS ONLY | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, UBIQUINOL CYTOCHROME C OXIDOREDUCTASE | | Authors: | Xia, D, Yu, C.A, Kim, H, Xia, J.Z, Kachurin, A, Zhang, L, Yu, L, Deisenhofer, J. | | Deposit date: | 1997-05-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria.
Science, 277, 1997
|
|
1HWT
 
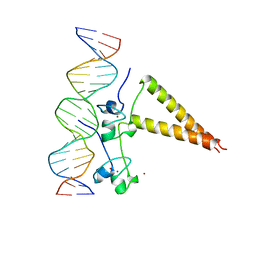 | | STRUCTURE OF A HAP1/DNA COMPLEX REVEALS DRAMATICALLY ASYMMETRIC DNA BINDING BY A HOMODIMERIC PROTEIN | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*C)-3'), PROTEIN (HEME ACTIVATOR PROTEIN), ... | | Authors: | King, D.A, Zhang, L, Guarente, L, Marmorstein, R. | | Deposit date: | 1998-09-17 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a HAP1-DNA complex reveals dramatically asymmetric DNA binding by a homodimeric protein.
Nat.Struct.Biol., 6, 1999
|
|
7XSR
 
 | | Structure of Craspase-target RNA | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSQ
 
 | | Structure of the Craspase | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
3B8U
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221A | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-11-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
8J8J
 
 | | Membrane bound PRTase, C3 symmetry, donor bound | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Decaprenyl-phosphate phosphoribosyltransferase, ... | | Authors: | Wu, F.Y, Gao, S, Zhang, L, Rao, Z.H. | | Deposit date: | 2023-05-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural analysis of phosphoribosyltransferase-mediated cell wall precursor synthesis in Mycobacterium tuberculosis.
Nat Microbiol, 9, 2024
|
|
2QFR
 
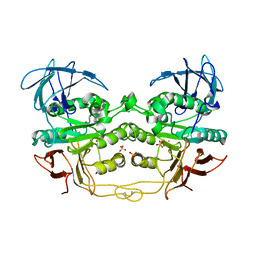 | | Crystal structure of red kidney bean purple acid phosphatase with bound sulfate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, ... | | Authors: | Guddat, L.W, Schenk, G, Gahan, L.R, Elliot, T.W, Leung, E. | | Deposit date: | 2007-06-27 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a purple acid phosphatase, representing different steps of this enzyme's catalytic cycle.
Bmc Struct.Biol., 8, 2008
|
|
7E6T
 
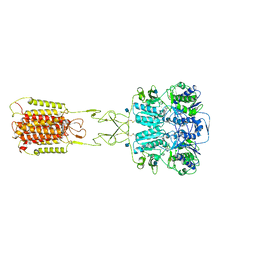 | | Structural insights into the activation of human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYCLOMETHYLTRYPTOPHAN, ... | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
7E6U
 
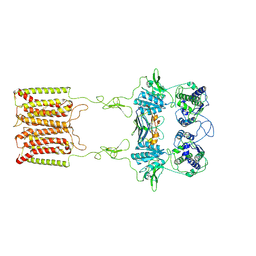 | | the complex of inactive CaSR and NB2D11 | | Descriptor: | Extracellular calcium-sensing receptor, NB-2D11 | | Authors: | Geng, Y, Chen, X.C, Wang, L, Cui, Q.Q, Ding, Z.Y, Han, L, Kou, Y.J, Zhang, W.Q, Wang, H.N, Jia, X.M, Dai, M, Shi, Z.Z, Li, Y.Y, Li, X.Y. | | Deposit date: | 2021-02-24 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural insights into the activation of human calcium-sensing receptor.
Elife, 10, 2021
|
|
3KBS
 
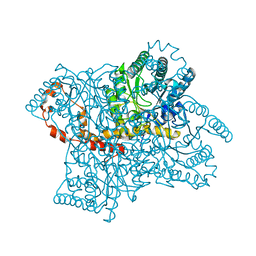 | |
3KBM
 
 | |
3KBV
 
 | |
3KBW
 
 | |
