8WCN
 
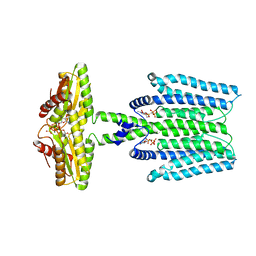 | | Cryo-EM structure of PAO1-ImcA with GMPCPP | | Descriptor: | Diguanylate cyclase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Zhan, X.L, Zhang, K, Wang, C.C, Fan, Q, Tang, X.J, Zhang, X, Wang, K, Fu, Y, Liang, H.H. | | Deposit date: | 2023-09-13 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A c-di-GMP signaling module controls responses to iron in Pseudomonas aeruginosa.
Nat Commun, 15, 2024
|
|
2ADU
 
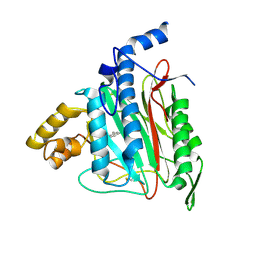 | | Human Methionine Aminopeptidase Complex with 4-Aryl-1,2,3-triazole Inhibitor | | Descriptor: | 4-(3-METHYLPHENYL)-1H-1,2,3-TRIAZOLE, COBALT (II) ION, Methionine aminopeptidase 2 | | Authors: | Kallander, L.S, Lu, Q, Chen, W, Tomaszek, T, Yang, G, Tew, D, Meek, T.D, Hofmann, G.A, Schulz-Pritchard, C.K, Smith, W.W, Janson, C.A, Ryan, M.D, Zhang, G.F, Johanson, K.O, Kirkpatrick, R.B, Ho, T.F, Fisher, P.W, Mattern, M.R, Johnson, R.K, Hansbury, M.J, Winkler, J.D, Ward, K.W, Veber, D.F, Thompson, S.K. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 4-Aryl-1,2,3-triazole: A Novel Template for a Reversible Methionine Aminopeptidase 2 Inhibitor, Optimized To Inhibit Angiogenesis in Vivo
J.Med.Chem., 48, 2005
|
|
8I09
 
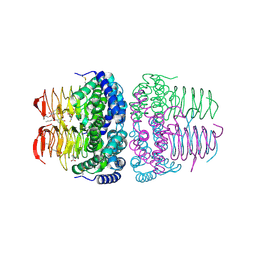 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with butyl gallate | | Descriptor: | CYSTEINE, PHOSPHATE ION, Serine acetyltransferase, ... | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
8I06
 
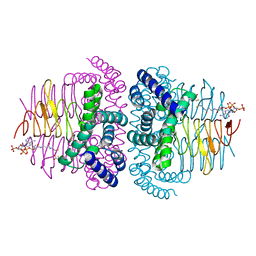 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with CoA | | Descriptor: | COENZYME A, CYSTEINE, Serine acetyltransferase | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
8I04
 
 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with serine | | Descriptor: | PHOSPHATE ION, SERINE, Serine acetyltransferase | | Authors: | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
7FD4
 
 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | Deposit date: | 2021-07-16 | | Release date: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
7FD5
 
 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | Deposit date: | 2021-07-16 | | Release date: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
6BDG
 
 | | HFQ monomer in spacegroup p6 at 1.93 angstrom resolution | | Descriptor: | RNA-binding protein Hfq | | Authors: | Brown, C, Zhang, K, Seo, C, Ellis, M.J, Hanniford, D.B, Junop, M. | | Deposit date: | 2017-10-23 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | HFQ monomer in spacegroup p6 at 1.93 angstrom resolution
To Be Published
|
|
7FID
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon proteasecomplex (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-07-31 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FIZ
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-08-01 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
7FIE
 
 | | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Su, S, Huang, K, Zhang, K, Chang, C.I. | | Deposit date: | 2021-07-31 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Processive cleavage of substrate at individual proteolytic active sites of the Lon protease complex.
Sci Adv, 7, 2021
|
|
5TG1
 
 | | 1.40 A resolution structure of Norovirus 3CL protease in complex with the a m-chlorophenyl substituted macrocyclic inhibitor (17-mer) | | Descriptor: | (4S,7S,17S)-17-(3-chlorophenyl)-7-(hydroxymethyl)-4-(2-methylpropyl)-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione, 3C-LIKE PROTEASE, CHLORIDE ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Kankanamalage, A.C.G, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis, and evaluation of a novel series of macrocyclic inhibitors of norovirus 3CL protease.
Eur J Med Chem, 127, 2016
|
|
6WLM
 
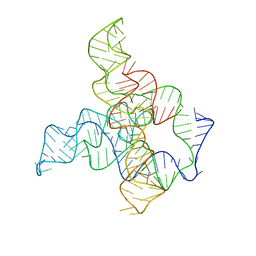 | | F. nucleatum glycine riboswitch with glycine models, 7.4 Angstrom resolution | | Descriptor: | RNA (171-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLR
 
 | | SAM-IV riboswitch with SAM models, 4.8 Angstrom resolution | | Descriptor: | RNA (119-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLS
 
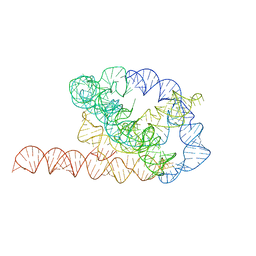 | | Tetrahymena ribozyme models, 6.8 Angstrom resolution | | Descriptor: | RNA (388-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
5TG2
 
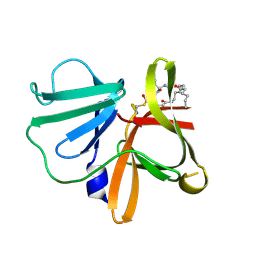 | | 1.75 A resolution structure of Norovirus 3CL protease in complex with the a n-pentyl substituted macrocyclic inhibitor (17-mer) | | Descriptor: | (4S,7S,17R)-7-(hydroxymethyl)-4-(2-methylpropyl)-17-pentyl-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Kankanamalage, A.C.G, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, and evaluation of a novel series of macrocyclic inhibitors of norovirus 3CL protease.
Eur J Med Chem, 127, 2016
|
|
6G6Q
 
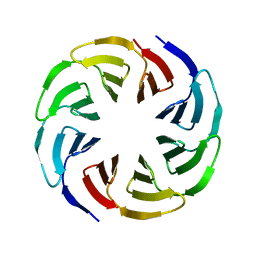 | | Crystal structure of the computationally designed Ika4 protein | | Descriptor: | Ika4 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
7T40
 
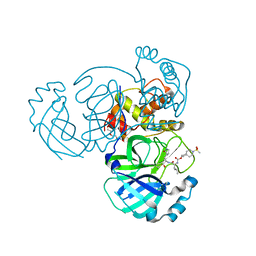 | | Structure of MERS 3CL protease in complex with inhibitor 10c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T42
 
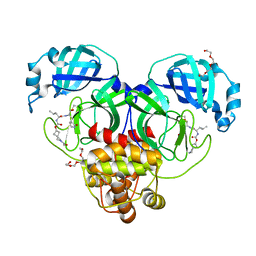 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 2c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[2-(2-methylpropanoyl)-2-azaspiro[3.3]heptan-6-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-[(N-{[(2-acetyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T49
 
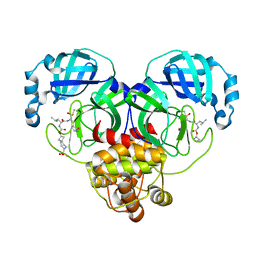 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 10c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(methanesulfonyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T3Y
 
 | | Structure of MERS 3CL protease in complex with inhibitor 8c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T4B
 
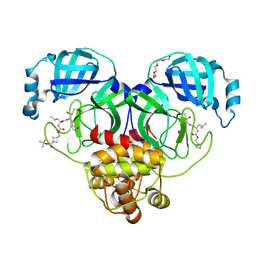 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T46
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 8c | | Descriptor: | (1R,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-{[N-({[7-(2-methylpropanoyl)-7-azaspiro[3.5]nonan-2-yl]oxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T41
 
 | | Structure of MERS 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[1-(tert-butoxycarbonyl)azetidin-3-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
7T43
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-[(N-{[(2-methyl-2-azaspiro[3.3]heptan-6-yl)oxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Chamandi, S.D, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-12-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease.
J.Med.Chem., 65, 2022
|
|
