5KPK
 
 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0209 | | Descriptor: | (4~{S})-3-cyclopropyl-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5VZ3
 
 | |
5VZ4
 
 | | Receptor-growth factor crystal structure at 2.20 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, GDNF family receptor alpha-like, ... | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2017-05-26 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15.
Nature, 550, 2017
|
|
1HYN
 
 | |
8KFT
 
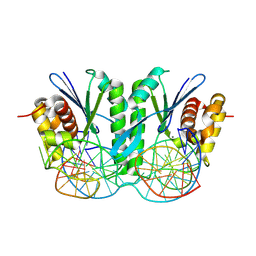 | | Crystal structure of ZmMOC1 in complex with a nicked Holliday junction soaked in Mn2+ for 15 seconds | | Descriptor: | DNA (25-MER), DNA (33-MER), DNA (5'-D(P*CP*AP*CP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
3JCD
 
 | | Structure of Escherichia coli EF4 in posttranslocational ribosomes (Post EF4) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
8KFU
 
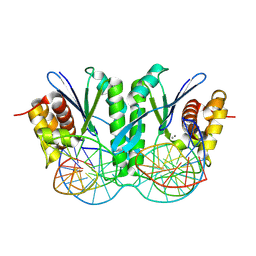 | | Crystal structure of ZmMOC1 in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFV
 
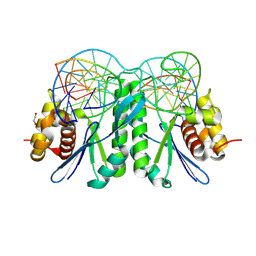 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 180 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (25-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFR
 
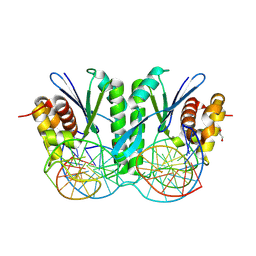 | | Crystal structure of ZmMOC1/nicked Holliday junction/Ca2+ complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (25-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFW
 
 | | Crystal structure of ZmMOC1 K229A in complex with a nicked Holliday junction soaked in Mn2+ for 600 seconds | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (33-MER), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8KFS
 
 | | Crystal structure of ZmMOC1/nicked Holliday junction complex at ground state | | Descriptor: | DNA (25-MER), DNA (33-MER), DNA (5'-D(P*CP*AP*CP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Zhang, D, Luo, Z, Lin, Z. | | Deposit date: | 2023-08-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | MOC1 cleaves Holliday junctions through a cooperative nick and counter-nick mechanism mediated by metal ions.
Nat Commun, 15, 2024
|
|
8P3S
 
 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
3JCE
 
 | | Structure of Escherichia coli EF4 in pretranslocational ribosomes (Pre EF4) | | Descriptor: | 16S ribosomal RNA, 23 ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, D, Yan, K, Liu, G, Song, G, Luo, J, Shi, Y, Cheng, E, Wu, S, Jiang, T, Low, J, Gao, N, Qin, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | EF4 disengages the peptidyl-tRNA CCA end and facilitates back-translocation on the 70S ribosome
Nat. Struct. Mol. Biol., 23, 2016
|
|
2JY5
 
 | | NMR structure of Ubiquilin 1 UBA domain | | Descriptor: | Ubiquilin-1 | | Authors: | Zhang, D, Raasi, S, Fushman, D. | | Deposit date: | 2007-12-06 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains
J.Mol.Biol., 377, 2008
|
|
2JY6
 
 | |
8P3V
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 3 | | Descriptor: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8PIV
 
 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-06-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3U
 
 | |
8P3T
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 1 | | Descriptor: | Glutamate receptor 1 flip isoform, Voltage-dependent calcium channel gamma-3 subunit | | Authors: | Zhang, D, Krieger, J, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3W
 
 | | Homomeric GluA1 in tandem with TARP gamma-3, desensitized conformation 4 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 1 flip isoform, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
8P3Q
 
 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor 2, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
1HTD
 
 | | STRUCTURAL INTERACTION OF NATURAL AND SYNTHETIC INHIBITORS WITH THE VENOM METALLOPROTEINASE, ATROLYSIN C (HT-D) | | Descriptor: | ATROLYSIN C, CALCIUM ION, ZINC ION | | Authors: | Zhang, D, Botos, I, Gomis-Rueth, F.-X, Doll, R, Blood, C, Njoroge, F.G, Fox, J.W, Bode, W, Meyer, E.F. | | Deposit date: | 1994-01-20 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural interaction of natural and synthetic inhibitors with the venom metalloproteinase, atrolysin C (form d).
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
6CZV
 
 | | BRD4(BD1) complexed with 2759 | | Descriptor: | 1,2-ETHANEDIOL, 1-benzyl-5-(3,5-dimethyl-1,2-oxazol-4-yl)pyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design and Characterization of Novel Covalent Bromodomain and Extra-Terminal Domain (BET) Inhibitors Targeting a Methionine.
J. Med. Chem., 61, 2018
|
|
3NS8
 
 | |
6CZU
 
 | | BRD4(BD1) complexed with 3219 | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-({4-[(1R)-1-hydroxyethyl]phenyl}methyl)pyridin-2(1H)-one, Bromodomain-containing protein 4, ... | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-26 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Design and Characterization of Novel Covalent Bromodomain and Extra-Terminal Domain (BET) Inhibitors Targeting a Methionine.
J. Med. Chem., 61, 2018
|
|
