3BHG
 
 | | Crystal structure of adenylosuccinate lyase from Legionella pneumophila | | Descriptor: | Adenylosuccinate lyase, GLYCEROL, SULFATE ION | | Authors: | Chang, C, Li, H, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of adenylosuccinate lyase from Legionella pneumophila.
To be Published
|
|
3BJN
 
 | | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae, targeted domain 79-240 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, putative | | Authors: | Chang, C, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae.
To be Published
|
|
3M6J
 
 | | Crystal structure of unknown function protein from Leptospirillum rubarum | | Descriptor: | CHLORIDE ION, uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-15 | | Release date: | 2010-03-31 | | Last modified: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of unknown function protein from Leptospirillum rubarum
To be Published
|
|
3KWP
 
 | | Crystal structure of putative methyltransferase from Lactobacillus brevis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Predicted methyltransferase | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-01 | | Release date: | 2009-12-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of putative methyltransferase from Lactobacillus brevis
To be Published
|
|
3B48
 
 | |
3E0K
 
 | |
4HVM
 
 | | Crystal structure of tallysomycin biosynthesis protein TlmII | | Descriptor: | SULFATE ION, TlmII | | Authors: | Chang, C, Bigelow, L, Bearden, J, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-21 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Crystal structure of tallysomycin biosynthesis protein TlmII
To be Published
|
|
3MT1
 
 | | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti | | Descriptor: | Putative carboxynorspermidine decarboxylase protein, SULFATE ION | | Authors: | Chang, C, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-30 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti
To be Published
|
|
3N3T
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase complex with cyclic di-gmp | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
3M0Z
 
 | | Crystal structure of putative aldolase from Klebsiella pneumoniae. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Chang, C, Rakowski, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of putative aldolase from Klebsiella pneumoniae.
To be Published
|
|
3MGL
 
 | |
3MAJ
 
 | | Crystal structure of putative DNA processing protein DprA from Rhodopseudomonas palustris CGA009 | | Descriptor: | DNA processing chain A, SULFATE ION | | Authors: | Chang, C, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-23 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of putative DNA processing protein DprA from Rhodopseudomonas palustris
To be Published
|
|
4HD1
 
 | |
3MXQ
 
 | |
3NET
 
 | |
8J8D
 
 | |
8J8T
 
 | |
5BU9
 
 | | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 | | Descriptor: | Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Chang, C, Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333
To Be Published
|
|
3BOQ
 
 | |
3FG8
 
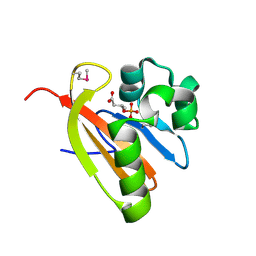 | | Crystal structure of PAS domain of RHA05790 | | Descriptor: | (3R)-3-(phosphonooxy)butanoic acid, uncharacterized protein RHA05790 | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-05 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PAS domain of RHA05790
To be Published
|
|
3N2Q
 
 | |
4EYQ
 
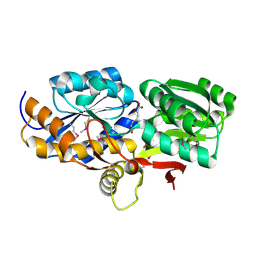 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 in complex with caffeic acid/3-(4-HYDROXY-PHENYL)PYRUVIC ACID | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, CAFFEIC ACID, Extracellular ligand-binding receptor | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3C0U
 
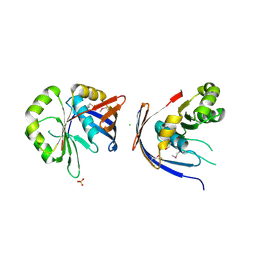 | | Crystal structure of E.coli yaeQ protein | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein yaeQ | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-21 | | Release date: | 2008-01-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of E.coli yaeQ protein.
To be Published
|
|
4EYK
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris BisB5 in complex with 3,4-dihydroxy benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 3,4-DIHYDROXYBENZOIC ACID, Twin-arginine translocation pathway signal | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
4F06
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 RPB_2270 in complex with P-HYDROXYBENZOIC ACID | | Descriptor: | Extracellular ligand-binding receptor, GLYCEROL, P-HYDROXYBENZOIC ACID, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-15 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
