7N40
 
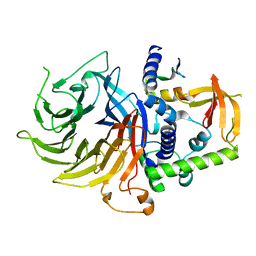 | | Crystal structure of LIN9-RbAp48-LIN37, a MuvB subcomplex | | Descriptor: | Histone-binding protein RBBP4, Isoform 2 of Protein lin-9 homolog, Protein lin-37 homolog | | Authors: | Asthana, A, Ramanan, P, Tripathi, S.M, Rubin, S.M. | | Deposit date: | 2021-06-02 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The MuvB complex binds and stabilizes nucleosomes downstream of the transcription start site of cell-cycle dependent genes.
Nat Commun, 13, 2022
|
|
7BJY
 
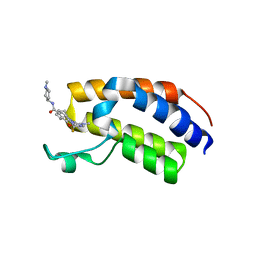 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to Ro3280 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Isoform 2 of Bromodomain testis-specific protein | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
9C87
 
 | |
9C88
 
 | |
5UOS
 
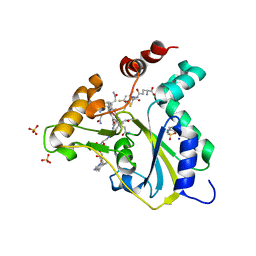 | | Crystal Structure of CblC (MMACHC) (1-238), a human B12 processing enzyme, complexed with an Antivitamin B12 | | Descriptor: | 1-ethynyl-2,4-difluorobenzene, 2-PHENYLAMINO-ETHANESULFONIC ACID, COBALAMIN, ... | | Authors: | Shanmuganathan, A, Karasik, A, Ruetz, M, Banerjee, R, Krautler, B, Koutmos, M. | | Deposit date: | 2017-02-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Antivitamin B12 Inhibition of the Human B12 -Processing Enzyme CblC: Crystal Structure of an Inactive Ternary Complex with Glutathione as the Cosubstrate.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1NJO
 
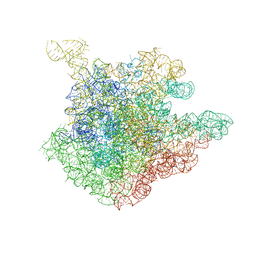 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a short substrate analog ACCPuromycin (ACCP) | | Descriptor: | 23S ribosomal RNA, RNA ACC(Puromycin) | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
1NJM
 
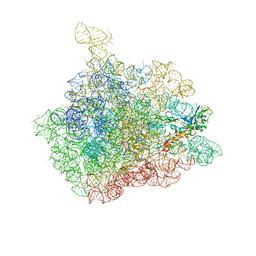 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a tRNA acceptor stem mimic (ASM) and the antibiotic sparsomycin | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, GENERAL STRESS PROTEIN CTC, ... | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
1NJP
 
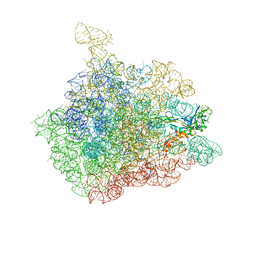 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a tRNA acceptor stem mimic (ASM) | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, GENERAL STRESS PROTEIN CTC, ... | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
6E5E
 
 | |
6E5Q
 
 | |
6E51
 
 | |
6E50
 
 | |
6E5R
 
 | |
6E7M
 
 | |
7MFY
 
 | |
7MFZ
 
 | |
7MFX
 
 | |
6E6L
 
 | |
6V1H
 
 | | Crystal structure of the bromodomain of human BRD7 bound to bromosporine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 7, Bromosporine | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1L
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to BI9564 | | Descriptor: | 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 4 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1E
 
 | | Crystal structure of the bromodomain of human BRD7 bound to BI7273 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-[(dimethylamino)methyl]-3,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 7 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-20 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6B8M
 
 | |
6MCV
 
 | |
6V1U
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to TP-472 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-acetylpyrrolo[1,2-a]pyrimidin-8-yl)-N-cyclopropyl-4-methylbenzamide, Bromodomain-containing protein 4 | | Authors: | Chan, A, Karim, M.R, Schonbrunn, E. | | Deposit date: | 2019-11-21 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6B8Q
 
 | |
