2AQY
 
 | |
5AB8
 
 | | High resolution X-ray structure of the N-terminal truncated form (residues 1-11) of Mycobacterium tuberculosis HbN | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Pesce, A, Bustamante, J.P, Bidon-Chanal, A, Boechi, L, Estrin, D.A, Luque, F.J, Sebilo, A, Guertin, M, Bolognesi, M, Ascenzi, P, Nardini, M. | | Deposit date: | 2015-08-04 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The N-Terminal Pre-A Region of Mycobacterium Tuberculosis 2/2Hbn Promotes No-Dioxygenase Activity.
FEBS J., 283, 2016
|
|
6XM6
 
 | | Crystal structure of cobalt-bound LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | COBALT (II) ION, Dioxygenase | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6OSW
 
 | | An order-to-disorder structural switch activates the FoxM1 transcription factor | | Descriptor: | Forkhead box M1 | | Authors: | Marceau, A.H, Rubin, S.M, Nerli, S, McShane, A.C, Sgourakis, N.G. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An order-to-disorder structural switch activates the FoxM1 transcription factor.
Elife, 8, 2019
|
|
7Q3H
 
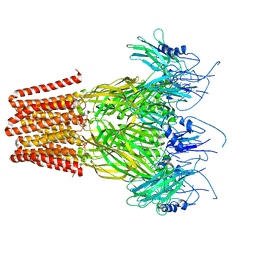 | | Pentameric ligand-gated ion channel, DeCLIC at pH 7 with 10 mM EDTA | | Descriptor: | Neur_chan_LBD domain-containing protein | | Authors: | Lycksell, M, Rovsnik, U, Hanke, A, Howard, R.J, Lindahl, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q3G
 
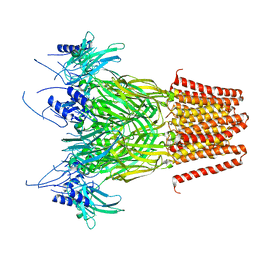 | | Pentameric ligand-gated ion channel, DeCLIC at pH 7 with 10 mM Ca2+ | | Descriptor: | CALCIUM ION, Neur_chan_LBD domain-containing protein | | Authors: | Licksell, M, Rovsnik, U, Hanke, A, Howard, R.J, Lindahl, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6MPP
 
 | |
4QMK
 
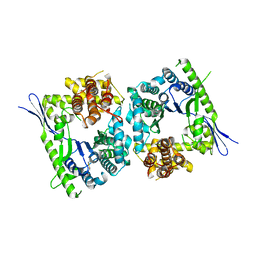 | | Crystal structure of type III effector protein ExoU (exoU) | | Descriptor: | BETA-MERCAPTOETHANOL, Type III secretion system effector protein ExoU | | Authors: | Halavaty, A.S, Tyson, G.H, Zhang, A, Hauser, A.R, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-16 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Phosphatidylinositol 4,5-Bisphosphate Binding Domain Mediates Plasma Membrane Localization of ExoU and Other Patatin-like Phospholipases.
J.Biol.Chem., 290, 2015
|
|
7QFU
 
 | | Crystal Structure of AtlA catalytic domain from Enterococcus feacalis | | Descriptor: | GLYCEROL, Peptidoglycan hydrolase | | Authors: | Zamboni, V, Barelier, S, Dixon, R, Galley, N, Ghanem, A, Cahuzac, H, Salamaga, B, Davis, P.J, Mesnage, S, Vincent, F. | | Deposit date: | 2021-12-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis for substrate recognition and septum cleavage by AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis.
J.Biol.Chem., 298, 2022
|
|
6FQ2
 
 | | Structure of minimal sequence for left -handed G-quadruplex formation | | Descriptor: | DNA (5'-D(*GP*TP*GP*GP*TP*GP*GP*TP*GP*GP*TP*G)-3'), POTASSIUM ION | | Authors: | Schmitt, E, Mechulam, Y, Phan, A.T, Heddi, B, Bakalar, B. | | Deposit date: | 2018-02-13 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Minimal Sequence for Left-Handed G-Quadruplex Formation.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6PQ3
 
 | | Crystal structure of GDP-bound KRAS with ten residues long internal tandem duplication in the switch II region | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dharmaiah, S, Chan, A.H, Tran, T.H, Simanshu, D.K. | | Deposit date: | 2019-07-08 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | RASinternal tandem duplication disrupts GTPase-activating protein (GAP) binding to activate oncogenic signaling.
J.Biol.Chem., 295, 2020
|
|
6GDH
 
 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*AP*CP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3') | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-23 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
6Q6R
 
 | | Recognition of different base tetrads by RHAU: X-ray crystal structure of G4 recognition motif bound to the 3-end tetrad of a DNA G-quadruplex | | Descriptor: | ATP-dependent DNA/RNA helicase DHX36, POTASSIUM ION, Parallel stranded DNA G-quadruplex | | Authors: | Heddi, B, Cheong, V.V, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2018-12-11 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of different base tetrads by RHAU (DHX36): X-ray crystal structure of the G4 recognition motif bound to the 3'-end tetrad of a DNA G-quadruplex.
J.Struct.Biol., 209, 2020
|
|
6FXC
 
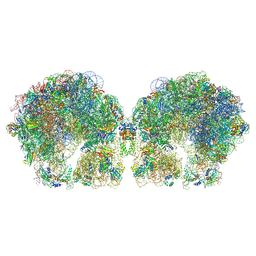 | | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Aibara, S, Zimmerman, E, Bashan, A, Kidmose, R, Amunts, A, Yonath, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-03-21 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (6.76 Å) | | Cite: | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus.
Nat Commun, 8, 2017
|
|
6GZ6
 
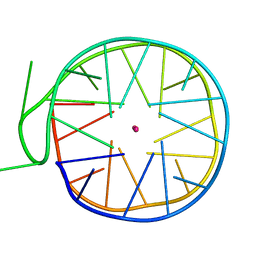 | | Structure of a left-handed G-quadruplex | | Descriptor: | DNA (27-MER), POTASSIUM ION | | Authors: | Bakalar, B, Heddi, B, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2018-07-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A Minimal Sequence for Left-Handed G-Quadruplex Formation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6GDN
 
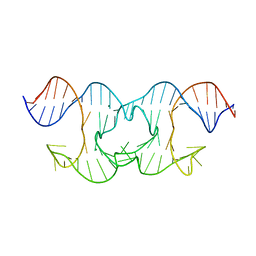 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | MAGNESIUM ION, Telomere DNA (42-MER) | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
6KVB
 
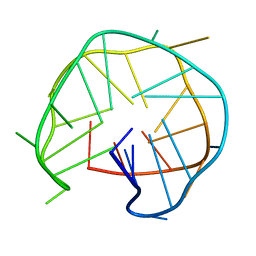 | | Structure of an intra-locked G-quadruplex | | Descriptor: | DNA (28-mer) | | Authors: | Maity, A, Winnerdy, F.R, Chang, W.D, Chen, G, Phan, A.T. | | Deposit date: | 2019-09-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Intra-locked G-quadruplex structures formed by irregular DNA G-rich motifs.
Nucleic Acids Res., 48, 2020
|
|
6E6I
 
 | | Crystal structure of 4-methyl HOPDA bound to LigY from Sphingobium sp. strain SYK-6 | | Descriptor: | (1Z,3E)-5-carboxy-3-methyl-5-oxo-1-phenylpenta-1,3-dien-1-olate, 2,2',3-trihydroxy-3'-methoxy-5,5'-dicarboxybiphenyl meta-cleavage compound hydrolase, ZINC ION | | Authors: | Kuatsjah, E, Chan, A.C, Hurst, T.E, Snieckus, V, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2018-07-24 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal- and Serine-Dependent Meta-Cleavage Product Hydrolases Utilize Similar Nucleophile-Activation Strategies
Acs Catalysis, 8, 2018
|
|
7ZLL
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with the 5-[(morpholin-4-yl)methyl]quinolin-8-ol inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(morpholin-4-ylmethyl)quinolin-8-ol, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Kantsadi, A.L, Chandran, A.V. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
8A98
 
 | | CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : snoRNA MUTANT | | Descriptor: | 40S ribosomal protein S12, 40S ribosomal protein S14, 40S ribosomal protein S19-like protein, ... | | Authors: | Rajan, K.S, Yonath, A, Bashan, A. | | Deposit date: | 2022-06-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural and mechanistic insights into the function of Leishmania ribosome lacking a single pseudouridine modification.
Cell Rep, 43, 2024
|
|
8A3W
 
 | |
6EZI
 
 | |
7L6D
 
 | | Crystal structure of the second bromodomain (BD2) of human BRD2 bound to bromosporine | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, Bromosporine, ... | | Authors: | Karim, M.R, Bikowitz, M.J, Chan, A, Schonbrunn, E. | | Deposit date: | 2020-12-23 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
6ETA
 
 | |
6GDS
 
 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | Telomeric DNA (5' CTAACCCTAA) 10mer, Telomeric DNA (5'-TTAGGGTTAG)-3') 10mer | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
