1L8W
 
 | | Crystal Structure of Lyme Disease Variable Surface Antigen VlsE of Borrelia burgdorferi | | Descriptor: | VlsE1 | | Authors: | Eicken, C, Sharma, V, Klabunde, T, Lawrenz, M.B, Hardham, J.M, Norris, S.J, Sacchettini, J.C. | | Deposit date: | 2002-03-21 | | Release date: | 2002-06-19 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Lyme disease variable surface antigen VlsE of Borrelia burgdorferi.
J.Biol.Chem., 277, 2002
|
|
1QAD
 
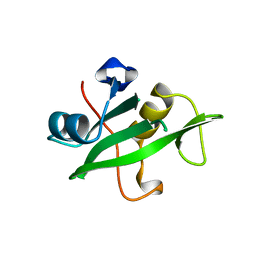 | | Crystal Structure of the C-Terminal SH2 Domain of the P85 alpha Regulatory Subunit of Phosphoinositide 3-Kinase: An SH2 domain mimicking its own substrate | | Descriptor: | PI3-KINASE P85 ALPHA SUBUNIT | | Authors: | Hoedemaeker, P.J, Siegal, G, Roe, M, Driscoll, P.C, Abrahams, J.P.A. | | Deposit date: | 1999-02-26 | | Release date: | 1999-10-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the C-terminal SH2 domain of the p85alpha regulatory subunit of phosphoinositide 3-kinase: an SH2 domain mimicking its own substrate.
J.Mol.Biol., 292, 1999
|
|
1W39
 
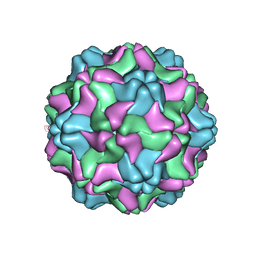 | | Crystal structure of an artificial top component of turnip yellow mosaic virus | | Descriptor: | TURNIP YELLOW MOSAIC VIRUS EMPTY CAPSID | | Authors: | van Roon, A.M.M, Bink, H.H.J, Plaisier, J.R, Pleij, C.W.A, Abrahams, J.P, Pannu, N.S. | | Deposit date: | 2004-07-14 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Crystal Structure of an Empty Capsid of Turnip Yellow Mosaic Virus.
J.Mol.Biol., 341, 2004
|
|
1A1Q
 
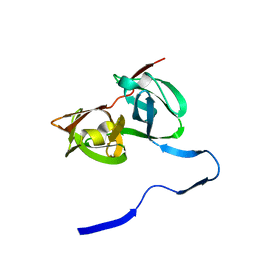 | | HEPATITIS C VIRUS NS3 PROTEINASE | | Descriptor: | NS3 PROTEINASE, ZINC ION | | Authors: | Love, R.A, Parge, H.E, Wickersham, J.A, Hostomsky, Z, Habuka, N, Moomaw, E.W, Adachi, T, Hostomska, Z. | | Deposit date: | 1997-12-12 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site.
Cell(Cambridge,Mass.), 87, 1996
|
|
1R49
 
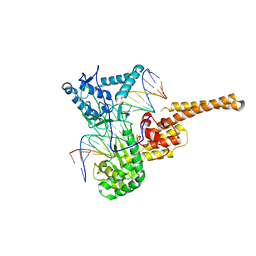 | | Human topoisomerase I (Topo70) double mutant K532R/Y723F | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(P*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3', DNA topoisomerase I | | Authors: | Interthal, H, Quigley, P.M, Hol, W.G, Champoux, J.J. | | Deposit date: | 2003-10-03 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | The role of lysine 532 in the catalytic mechanism of human topoisomerase I.
J.Biol.Chem., 279, 2004
|
|
1TTB
 
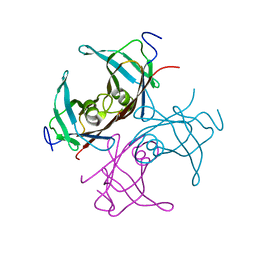 | |
3OOS
 
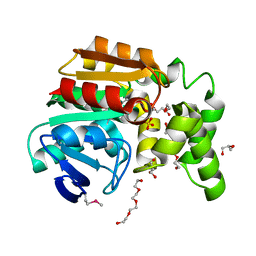 | | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne | | Descriptor: | Alpha/beta hydrolase family protein, GLYCEROL, SULFATE ION, ... | | Authors: | Fan, Y, Tan, K, Bigelow, L, Hamilton, J, Li, H, Zhou, Y, Clancy, S, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne
To be Published
|
|
1EAL
 
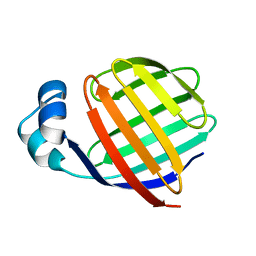 | | NMR STUDY OF ILEAL LIPID BINDING PROTEIN | | Descriptor: | ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Rueterjans, H, Hamilton, J.A, Sacchettini, J.C. | | Deposit date: | 1996-08-28 | | Release date: | 1997-01-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Flexibility is a likely determinant of binding specificity in the case of ileal lipid binding protein.
Structure, 4, 1996
|
|
2M72
 
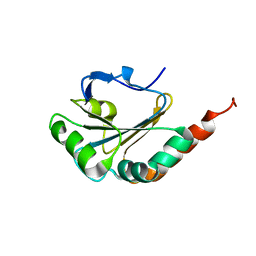 | | Solution structure of uncharacterized thioredoxin-like protein PG_2175 from Porphyromonas gingivalis | | Descriptor: | Uncharacterized thioredoxin-like protein | | Authors: | Harris, R, Ahmed, M, Attonito, J, Bonanno, J.B, Chamala, S, Chowdhury, S, Evans, B, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Seidel, R.D, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-16 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of uncharacterized thioredoxin-like protein PG_2175 from Porphyromonas gingivalis
To be Published
|
|
2M71
 
 | | Solution structure of the a C-terminal domain of translation initiation factor IF-3 from Campylobacter jejuni | | Descriptor: | Translation initiation factor IF-3 | | Authors: | Harris, R, Ahmed, M, Attonito, J, Bonanno, J.B, Chamala, S, Chowdhury, S, Evans, B, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Seidel, R.D, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-16 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the a C-terminal domain of translation initiation factor IF-3 from Campylobacter jejuni
To be Published
|
|
1E04
 
 | | PLASMA BETA ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, GLYCEROL, ... | | Authors: | Mccoy, A.J, Skinner, R, Abrahams, J.-P, Pei, X.Y, Carrell, R.W. | | Deposit date: | 2000-03-09 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Beta-Antithrombin and the Effect of Glycosylation on Antithrombin'S Heparin Affinity and Activity.
J.Mol.Biol., 326, 2003
|
|
1AZX
 
 | | ANTITHROMBIN/PENTASACCHARIDE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4-di-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-3-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2,3,6-tri-O-sulfo-alpha-D-glucopyranoside, ANTITHROMBIN | | Authors: | Jin, L, Abrahams, J.P, Skinner, R, Petitou, M, Pike, R.N, Carrell, R.W. | | Deposit date: | 1997-11-23 | | Release date: | 1999-01-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The anticoagulant activation of antithrombin by heparin.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1E03
 
 | | PLASMA ALPHA ANTITHROMBIN-III AND PENTASACCHARIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4-di-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-3-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2,3,6-tri-O-sulfo-alpha-D-glucopyranoside, ... | | Authors: | McCoy, A.J, Jin, L, Abrahams, J.-P, Skinner, R, Carrell, R.W. | | Deposit date: | 2000-03-09 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Beta-Antithrombin and the Effect of Glycosylation on Antithrombin'S Heparin Affinity and Activity.
J.Mol.Biol., 326, 2003
|
|
1E05
 
 | | PLASMA ALPHA ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | McCoy, A.J, Skinner, R, Abrahams, J.-P, Pei, X.Y, Carrell, R.W. | | Deposit date: | 2000-03-09 | | Release date: | 2000-06-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of Beta-Antithrombin and the Effect of Glycosylation on Antithrombin'S Heparin Affinity and Activity.
J.Mol.Biol., 326, 2003
|
|
2CJI
 
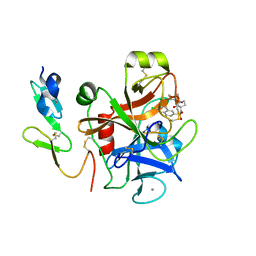 | | Crystal structure of a Human Factor Xa inhibitor complex | | Descriptor: | 6-CHLORO-N-{(3S)-1-[(1S)-1-METHYL-2-(4-MORPHOLINYL)-2-OXO ETHYL]-2-OXO-3-PYRROLIDINYL}-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Watson, N.S, Campbell, M, Chan, C, Convery, M.A, Hamblin, J.N, Kelly, H.A, King, N.P, Mason, A.M, Mitchell, C, Patel, V.K, Senger, S, Shah, G.P, Weston, H.E, Whitworth, C, Young, R.J. | | Deposit date: | 2006-04-03 | | Release date: | 2006-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and Synthesis of Orally Active Pyrrolidin-2-One-Based Factor Xa Inhibitors
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2M4M
 
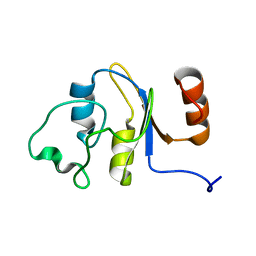 | | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata | | Descriptor: | hypothetical protein | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Patel, H, Seidel, R.D, Chook, Y.M, Rout, M.P, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata
To be Published
|
|
2M4N
 
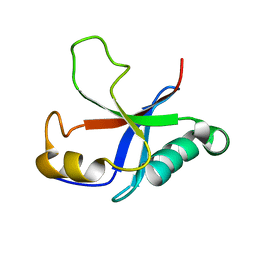 | | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans | | Descriptor: | Protein AFD-1, isoform a | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Seidel, R.D, Liddington, R.C, Weis, W.I, Nelson, W.J, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Assembly, Dynamics and Evolution of Cell-Cell and Cell-Matrix Adhesions (CELLMAT) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the putative Ras interaction domain of AFD-1, isoform a from Caenorhabditis elegans
To be Published
|
|
1VR2
 
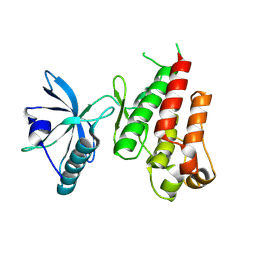 | | HUMAN VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 (KDR) KINASE DOMAIN | | Descriptor: | PROTEIN (VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR KINASE) | | Authors: | Mctigue, M, Wickersham, J, Pinko, C, Showalter, R, Parast, C, Tempczyk-Russell, A, Gehring, M, Mroczkowski, B, Kan, C, Villafranca, J, Appelt, K. | | Deposit date: | 1998-12-03 | | Release date: | 2000-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the kinase domain of human vascular endothelial growth factor receptor 2: a key enzyme in angiogenesis.
Structure Fold.Des., 7, 1999
|
|
1ONV
 
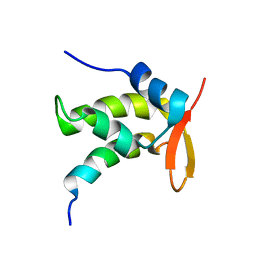 | | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNAP II CTD Phosphatase FCP1 | | Descriptor: | Transcription initiation factor IIF, alpha subunit, serine phosphatase FCP1a | | Authors: | Nguyen, B.D, Abbott, K.L, Potempa, K, Kobor, M.S, Archambault, J, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNA polymerase II carboxyl-terminal domain phosphatase FCP1
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1JY1
 
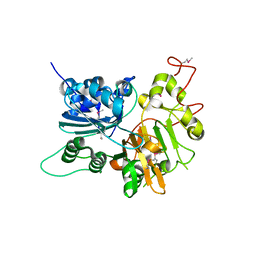 | | CRYSTAL STRUCTURE OF HUMAN TYROSYL-DNA PHOSPHODIESTERASE (TDP1) | | Descriptor: | TYROSYL-DNA PHOSPHODIESTERASE | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 2001-09-10 | | Release date: | 2002-02-20 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1.
Structure, 10, 2002
|
|
1UZ8
 
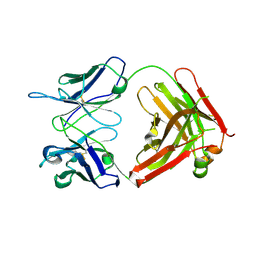 | | anti-Lewis X Fab fragment in complex with Lewis X | | Descriptor: | IGG FAB (IGG3, KAPPA) HEAVY CHAIN 291-2G3-A, KAPPA) LIGHT CHAIN 291-2G3-A, ... | | Authors: | Van Roon, A.M.M, Pannu, N.S, De Vrind, J.P.M, Hokke, C.H, Deelder, A.M, Van Der marel, G.A, Van Boom, J.H, Abrahams, J.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of an Anti-Lewis X Fab Fragment in Complex with its Lewis X Antigen
Structure, 12, 2004
|
|
2VXH
 
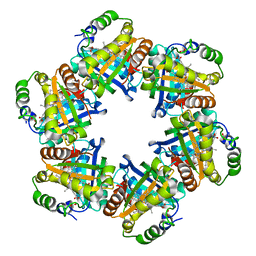 | | The crystal structure of chlorite dismutase: a detox enzyme producing molecular oxygen | | Descriptor: | CARBONATE ION, CHLORITE DISMUTASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | De Geus, D.C, Thomassen, E.A.J, Hagedoorn, P.L, Pannu, N.S, Abrahams, J.P. | | Deposit date: | 2008-07-04 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Chlorite Dismutase, a Detoxifying Enzyme Producing Molecular Oxygen
J.Mol.Biol., 387, 2009
|
|
1EIO
 
 | | ILEAL LIPID BINDING PROTEIN IN COMPLEX WITH GLYCOCHOLATE | | Descriptor: | GLYCOCHOLIC ACID, ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Hamilton, J.A, Sacchettini, J.C, Rueterjans, H. | | Deposit date: | 2000-02-27 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ileal lipid binding protein in complex with glycocholate.
Eur.J.Biochem., 267, 2000
|
|
1SBS
 
 | | CRYSTAL STRUCTURE OF AN ANTI-HCG FAB | | Descriptor: | MONOCLONAL ANTIBODY 3A2, SULFATE ION | | Authors: | Fotinou, C, Beauchamp, J, Emsley, P, Dehaan, A, Schielen, W.J.G, Bos, E, Isaacs, N.W. | | Deposit date: | 1998-04-08 | | Release date: | 1999-04-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an Fab fragment against a C-terminal peptide of hCG at 2.0 A resolution.
J.Biol.Chem., 273, 1998
|
|
1UZ6
 
 | | anti-Lewis X Fab fragment uncomplexed | | Descriptor: | IGG FAB (IGG3, KAPPA) HEAVY CHAIN 291-2G3-A, KAPPA) LIGHT CHAIN 291-2G3-A, ... | | Authors: | Van Roon, A.M.M, Pannu, N.S, De Vrind, J.P.M, Hokke, C.H, Deelder, A.M, Van Der marel, G.A, Van Boom, J.H, Abrahams, J.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of an Anti-Lewis X Fab Fragment in Complex with its Lewis X Antigen
Structure, 12, 2004
|
|
