6ZHJ
 
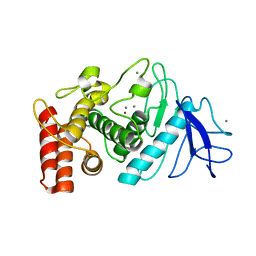 | | 3D electron diffraction structure of thermolysin from Bacillus thermoproteolyticus | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.26 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZHN
 
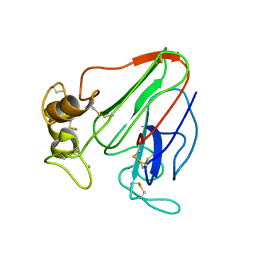 | | 3D electron diffraction structure of thaumatin from Thaumatococcus daniellii | | Descriptor: | CHLORIDE ION, Thaumatin-1 | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-23 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.76 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5UIY
 
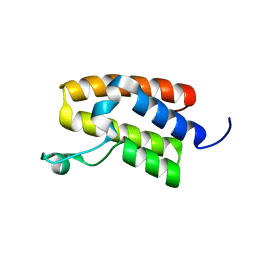 | | Structure of Bromodomain from human BAZ1A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bromodomain adjacent to zinc finger domain protein 1A | | Authors: | Oppikofer, M, Sudhamsu, J. | | Deposit date: | 2017-01-16 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | Non-canonical reader modules of BAZ1A promote recovery from DNA damage.
Nat Commun, 8, 2017
|
|
5N88
 
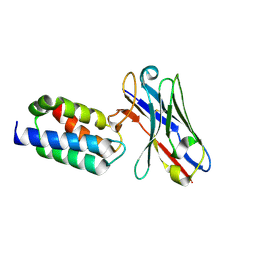 | | Crystal structure of antibody bound to viral protein | | Descriptor: | PC4 and SFRS1-interacting protein, VH59 antibody | | Authors: | Bao, L, Hannon, C, Cruz-Migoni, A, Ptchelkine, D, Sun, M.-y, Derveni, M, Bunjobpol, W, Chambers, J.S, Simmons, A, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intracellular immunization against HIV infection with an intracellular antibody that mimics HIV integrase binding to the cellular LEDGF protein.
Sci Rep, 7, 2017
|
|
6ZHB
 
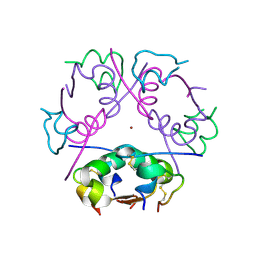 | | 3D electron diffraction structure of bovine insulin | | Descriptor: | Insulin, ZINC ION | | Authors: | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G, Ling, W.L, Abrahams, J.P. | | Deposit date: | 2020-06-22 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.25 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4WJI
 
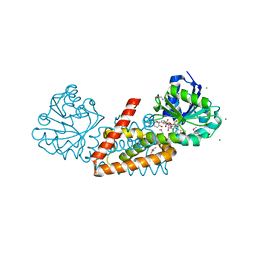 | | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP and tyrosine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shabalin, I.G, Cooper, D.R, Hou, J, Zimmerman, M.D, Stead, M, Hillerich, B.S, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-30 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP
to be published
|
|
4JN8
 
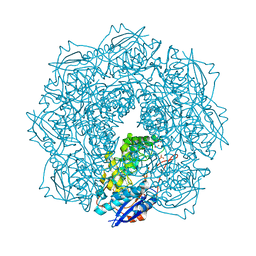 | | Crystal structure of an enolase (putative galactarate dehydratase, target efi-500740) from agrobacterium radiobacter, bound sulfate, no metal ion, ordered active site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ENOLASE, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Bouvier, J.T, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase (putative galactarate dehydratase, target efi-500740) from agrobacterium radiobacter, bound sulfate, no metal ion, ordered active site
To be Published
|
|
5A8Z
 
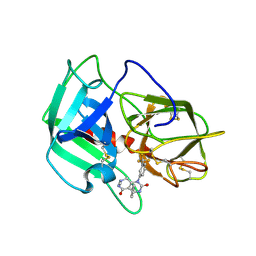 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(4R)-7-methyl-2,5-bis(oxidanylidene)-1-[3-(trifluoromethyl)phenyl]-3,4,6,8-tetrahydropyrimido[4,5-d]pyridazin-4-yl]benzenecarbonitrile, Neutrophil elastase, ... | | Authors: | von Nussbaum, F, Li, V.M, Meibom, D, Anlauf, S, Bechem, M, Delbeck, M, Gerisch, M, Harrenga, A, Karthaus, D, Lang, D, Lustig, K, Mittendorf, J, Schaefer, M, Schaefer, S, Schamberger, J. | | Deposit date: | 2015-07-17 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective Human Neutrophil Elastase Inhibitors with Novel Equatorial Ring Topology: in vivo Efficacy of the Polar Pyrimidopyridazine BAY-8040 in a Pulmonary Arterial Hypertension Rat Model.
ChemMedChem, 11, 2016
|
|
4JN7
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Bouvier, J.T, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE
To be Published
|
|
4Z44
 
 | | F454K Mutant of Tryptophan 7-halogenase PrnA | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase PrnA, ... | | Authors: | Shepherd, S.A, Karthikeyan, C, Latham, J, Struck, A.-W, Thompson, M.L, Menon, B, Levy, C.W, Leys, D, Micklefield, J. | | Deposit date: | 2015-04-01 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Extending the biocatalytic scope of regiocomplementary flavin-dependent halogenase enzymes.
Chem Sci, 6, 2015
|
|
2YOG
 
 | | Plasmodium falciparum thymidylate kinase in complex with a (thio)urea- alpha-deoxythymidine inhibitor | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-[[(2R,3S)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxol an-2-yl]methyl]thiourea, THYMIDYLATE KINASE | | Authors: | Huaqing, C, Carrero-Lerida, J, Silva, A.P.G, Whittingham, J.L, Brannigan, J.A, Ruiz-Perez, L.M, Read, K.D, Wilson, K.S, Gonzalez-Pacanowska, D, Gilbert, I.H. | | Deposit date: | 2012-10-24 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and Evaluation of Alpha-Thymidine Analogues as Novel Antimalarials.
J.Med.Chem., 55, 2012
|
|
4H83
 
 | | Crystal structure of Mandelate racemase/muconate lactonizing enzyme (EFI target:502127) | | Descriptor: | BICARBONATE ION, GLYCEROL, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-21 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing enzyme
To be Published
|
|
4CAS
 
 | | Serial femtosecond crystallography structure of a photosynthetic reaction center | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL A, ... | | Authors: | Johansson, L.C, Arnlund, D, Katona, G, White, T.A, Barty, A, DePonte, D.P, Shoeman, R.L, Wickstrand, C, Sharma, A, Williams, G.J, Aquila, A, Bogan, M.J, Caleman, C, Davidsson, J, Doak, R.B, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Kassemeyer, S, Kirian, R.A, Kupitz, C, Liang, M, Lomb, L, Malmerberg, E, Martin, A.V, Messerschmidt, M, Nass, K, Redecke, L, Seibert, M.M, Sjohamn, J, Steinbrener, J, Stellato, F, Wang, D, Wahlgren, W.Y, Weierstall, U, Westenhoff, S, Zatsepin, N.A, Boutet, S, Spence, J.C.H, Schlichting, I, Chapman, H.N, Fromme, P, Neutze, R. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography.
Nat Commun, 4, 2013
|
|
4YYC
 
 | | Crystal structure of trimethylamine methyltransferase from Sinorhizobium meliloti in complex with unknown ligand | | Descriptor: | CHLORIDE ION, Putative trimethylamine methyltransferase, UNKNOWN LIGAND | | Authors: | Shabalin, I.G, Porebski, P.J, Gasiorowska, O.A, Handing, K.B, Niedzialkowska, E, Cymborowski, M.T, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
6DEU
 
 | | Human caspase-6 A109T | | Descriptor: | Caspase-6 | | Authors: | Tubeleviciute-Aydin, A, Beautrait, A, Lynham, J, Sharma, G, Gorelik, A, Deny, L.J, Soya, N, Lukacs, G.L, Nagar, B, Marinier, A, LeBlanc, A.C. | | Deposit date: | 2018-05-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Identification of Allosteric Inhibitors against Active Caspase-6.
Sci Rep, 9, 2019
|
|
4MIJ
 
 | | Crystal structure of a Trap periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Galacturonate, space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-31 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
5W1M
 
 | | MACV GP1 CR1-07 Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR1-07 Fab heavy chain, ... | | Authors: | Raymond, D.D, Clark, L.E, Abraham, J. | | Deposit date: | 2017-06-03 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | Vaccine-elicited receptor-binding site antibodies neutralize two New World hemorrhagic fever arenaviruses.
Nat Commun, 9, 2018
|
|
6DEV
 
 | | Human caspase-6 E35K | | Descriptor: | Caspase-6 | | Authors: | Tubeleviciute-Aydin, A, Beautrait, A, Lynham, J, Sharma, G, Gorelik, A, Deny, L.J, Soya, N, Lukacs, G.L, Nagar, B, Marinier, A, LeBlanc, A.C. | | Deposit date: | 2018-05-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Identification of Allosteric Inhibitors against Active Caspase-6.
Sci Rep, 9, 2019
|
|
3ZBX
 
 | | X-ray Structure of c-Met kinase in complex with inhibitor 6-((6-(4- fluorophenyl)-(1,2,4)triazolo(4,3-b)(1,2,4)triazin-3-yl)methyl) quinoline. | | Descriptor: | 6-[[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]quinoline, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Wickersham, J, Ryan, K. | | Deposit date: | 2012-11-13 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lessons from (S)-6-(1-(6-(1-Methyl-1H-Pyrazol-4-Yl)-[1,2, 4]Triazolo[4,3-B]Pyridazin-3-Yl)Ethyl)Quinoline (Pf-04254644), an Inhibitor of Receptor Tyrosine Kinase C-met with High Protein Kinase Selectivity But Broad Phosphodiesterase Family Inhibition Leading to Myocardial Degeneration in Rats.
J.Med.Chem., 56, 2013
|
|
4N4U
 
 | | Crystal structure of ABC transporter solute binding protein BB0719 from Bordetella bronchiseptica RB50, TARGET EFI-510049 | | Descriptor: | GLYCEROL, Putative ABC transporter periplasmic solute-binding protein | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4J2F
 
 | | Crystal structure of a glutathione transferase family member from Ricinus communis, target EFI-501866 | | Descriptor: | GLYCEROL, Glutathione s-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-02-04 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Ricinus communis, target EFI-501866
TO BE PUBLISHED
|
|
4KDY
 
 | | Crystal structure of maleylacetoacetate isomerase from Anaeromyxobacter dehalogenans 2CP-1, Target EFI-507175, with bound GSH in the active site | | Descriptor: | CITRIC ACID, GLUTATHIONE, GLYCEROL, ... | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-25 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of maleylacetoacetate isomerase from Anaeromyxobacter dehalogenans 2CP-1, Target EFI-507175, with bound GSH in the active site
To be Published
|
|
4KAE
 
 | | Crystal structure of Maleylacetoacetate isomerase from Anaeromyxobacter dehalogenans 2CP-1, TARGET EFI-507175, with bound dicarboxyethyl glutathione and citrate in the active site | | Descriptor: | CITRIC ACID, GAMMA-GLUTAMYL-S-(1,2-DICARBOXYETHYL)CYSTEINYLGLYCINE, GLYCEROL, ... | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Maleylacetoacetate isomerase from Anaeromyxobacter dehalogenans 2CP-1, TARGET EFI-507175, with bound dicarboxyethyl glutathione and citrate in the active site
To be Published
|
|
4HI7
 
 | | Crystal structure of glutathione transferase homolog from drosophilia mojavensis, TARGET EFI-501819, with bound glutathione | | Descriptor: | GI20122, GLUTATHIONE | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-10-11 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of glutathione transferase homolog from drosophilia mojavensis, TARGET EFI-501819, with bound glutathione
To be Published
|
|
4HNL
 
 | | Crystal structure of ENOLASE EGBG_01401 (TARGET EFI-502226) from Enterococcus gallinarum EG2 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N.F, Stead, M, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-10-19 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of ENOLASE EGBG_01401 from Enterococcus gallinarum EG2
To be Published
|
|
