1FXL
 
 | |
3K61
 
 | | Crystal structure of FBF-2/fog-1 FBEa complex | | Descriptor: | 5'-R(*UP*GP*UP*AP*AP*AP*AP*UP*C)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Opperman, L, Wickens, M, Hall, T.M.T. | | Deposit date: | 2009-10-08 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8DGA
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IV | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DG7
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex III | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DFV
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIa | | Descriptor: | CALCIUM ION, Endoribonuclease Dcr-1, Loquacious, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-22 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DGJ
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ib | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DG5
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex IIb | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B, ... | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
8DGI
 
 | | Structural Basis of MicroRNA Biogenesis by Dicer-1 and Its Partner Protein Loqs-PB - complex Ia | | Descriptor: | Endoribonuclease Dcr-1, Loquacious, isoform B | | Authors: | Jouravleva, K, Golovenko, D, Demo, G, Dutcher, R.C, Tanaka Hall, T.M, Zamore, P.D, Korostelev, A.A. | | Deposit date: | 2022-06-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis of microRNA biogenesis by Dicer-1 and its partner protein Loqs-PB.
Mol.Cell, 82, 2022
|
|
3BWT
 
 | |
3BX2
 
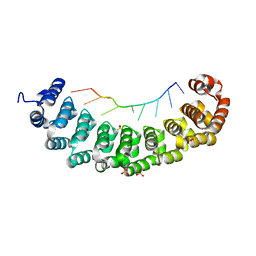 | | Puf4 RNA binding domain bound to HO endonuclease RNA 3' UTR recognition sequence | | Descriptor: | HO endonuclease 3' UTR binding sequence, Protein PUF4, SODIUM ION, ... | | Authors: | Miller, M.T, Higgin, J.J, Hall, T.M.T. | | Deposit date: | 2008-01-11 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p
Nat.Struct.Mol.Biol., 15, 2008
|
|
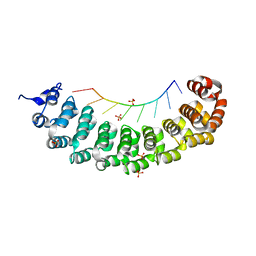
3BX3
 
 | |
7S02
 
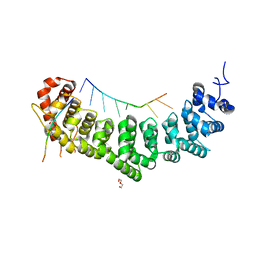 | |
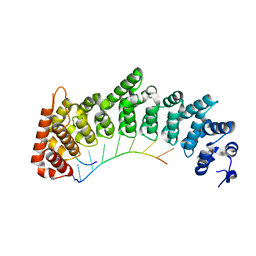
7RZZ
 
 | |
2AB5
 
 | | bI3 LAGLIDADG Maturase | | Descriptor: | SULFATE ION, mRNA maturase | | Authors: | Longo, A, Leonard, C.W, Bassi, G.S, Berndt, D, Krahn, J.M, Hall, T.M, Weeks, K.M. | | Deposit date: | 2005-07-14 | | Release date: | 2005-08-30 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution from DNA to RNA recognition by the bI3 LAGLIDADG maturase
Nat.Struct.Mol.Biol., 12, 2005
|
|
3T9E
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP, 5-(PP)-IP5 (5-IP7) and MgF3 (transition state mimic) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-DIPHOSPHATE, Inositol Pyrophosphate Kinase, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T9B
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP at pH 5.2 | | Descriptor: | Inositol Pyrophosphate Kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T54
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ATP and Cadmium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CADMIUM ION, Inositol Pyrophosphate Kinase | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-07-26 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T7A
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP at pH 5.2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-07-29 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T9F
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and 1,5-(PP)2-IP4 (1,5-IP8) | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], ADENOSINE-5'-DIPHOSPHATE, CADMIUM ION, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T9C
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP and inositol hexakisphosphate (IP6) | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Inositol Pyrophosphate Kinase, MAGNESIUM ION, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T9D
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP and 5-(PP)-IP5 (5-IP7) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, Inositol Pyrophosphate Kinase, MAGNESIUM ION, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T9A
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP at pH 7.0 | | Descriptor: | CADMIUM ION, Inositol Pyrophosphate Kinase, MAGNESIUM ION, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T99
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and in the absence of cadmium at pH 7.0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol Pyrophosphate Kinase, MAGNESIUM ION | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3V6Y
 
 | | crystal structure of FBF-2 in complex with a mutant gld-1 FBEa13 RNA | | Descriptor: | Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*AP*CP*UP*GP*UP*GP*CP*CP*AP*UP*AP*C)-3') | | Authors: | Qiu, C, Kershner, A, Wang, Y, Holley, C.H, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
3V71
 
 | | Crystal structure of PUF-6 in complex with 5BE13 RNA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Puf (Pumilio/fbf) domain-containing protein 7, confirmed by transcript evidence, ... | | Authors: | Qiu, C, Kershner, A, Wang, Y, Holley, C.H, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
