4E8X
 
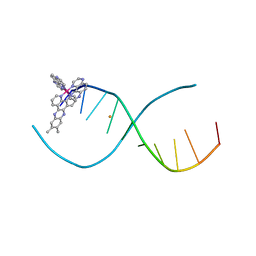 | | Lambda-[Ru(TAP)2(dppz-(Me)2)]2+ bound to d(CCGGCGCCGG)2 | | Descriptor: | (11,12-dimethyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium(2+), 5'-D(*CP*CP*GP*GP*CP*GP*CP*CP*GP*G)-3', BARIUM ION | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The effects of disubstitution on the binding of ruthenium complexes to DNA
Thesis, University of Reading, 2014
|
|
4YY2
 
 | |
6XR1
 
 | |
6XR2
 
 | |
4YIS
 
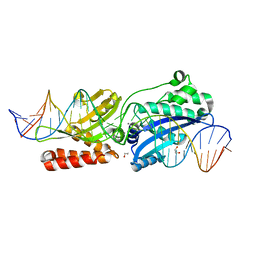 | | Crystal Structure of LAGLIDADG Meganuclease I-CpaMI Bound to Uncleaved DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (28-MER), ... | | Authors: | Hallinan, J.P, Kaiser, B.K, Stoddard, B.L. | | Deposit date: | 2015-03-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Indirect DNA Sequence Recognition and Its Impact on Nuclease Cleavage Activity.
Structure, 24, 2016
|
|
4YIT
 
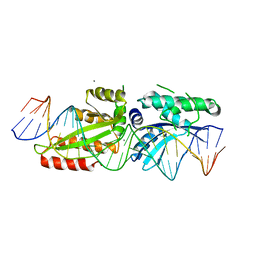 | |
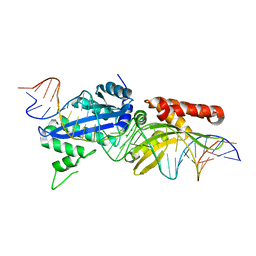
4YHX
 
 | |
4Z1Z
 
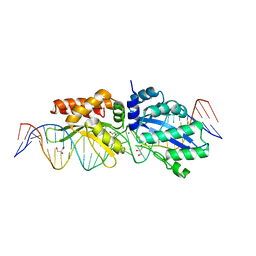 | |
4Z20
 
 | |
5ESP
 
 | |
5THG
 
 | |
4YY5
 
 | |
8ASM
 
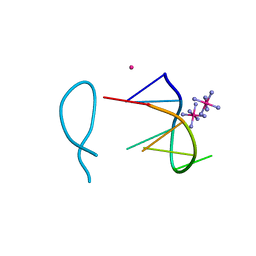 | | Cobalt(II) bound to a non-canonical quadruplex | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3') | | Authors: | Lambert, M.C, Hall, J.P. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identifying metal-DNA binding sites, what is the best method to get transition metals into a crystal system?
To Be Published
|
|
6HWG
 
 | |
8BAG
 
 | |
8BAE
 
 | |
8BAF
 
 | |
8OE3
 
 | |
8OE9
 
 | |
8OEC
 
 | |
8OEB
 
 | |
8OEA
 
 | |
8OE8
 
 | |
8OE7
 
 | |
8OEX
 
 | |
